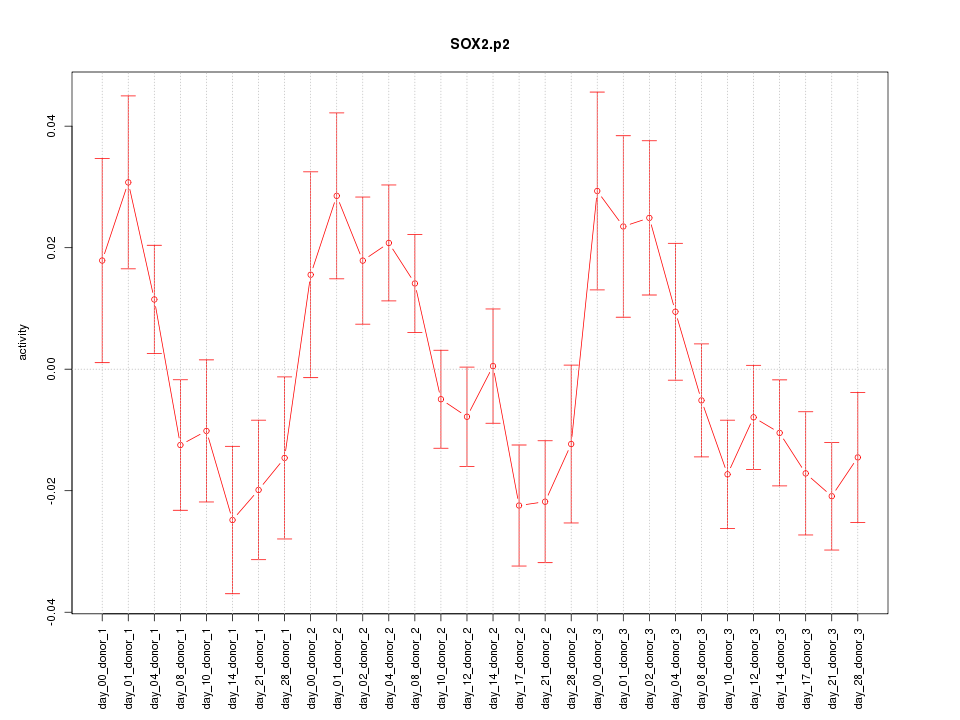
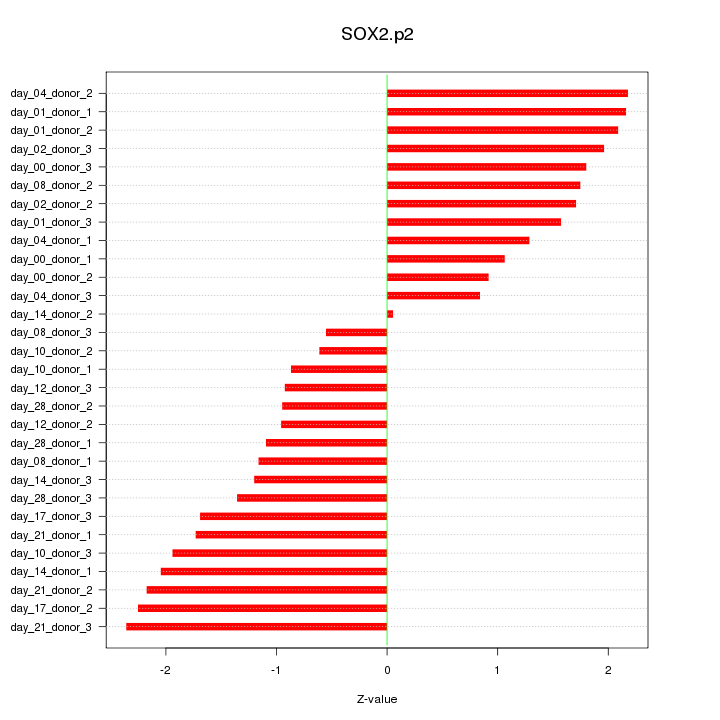
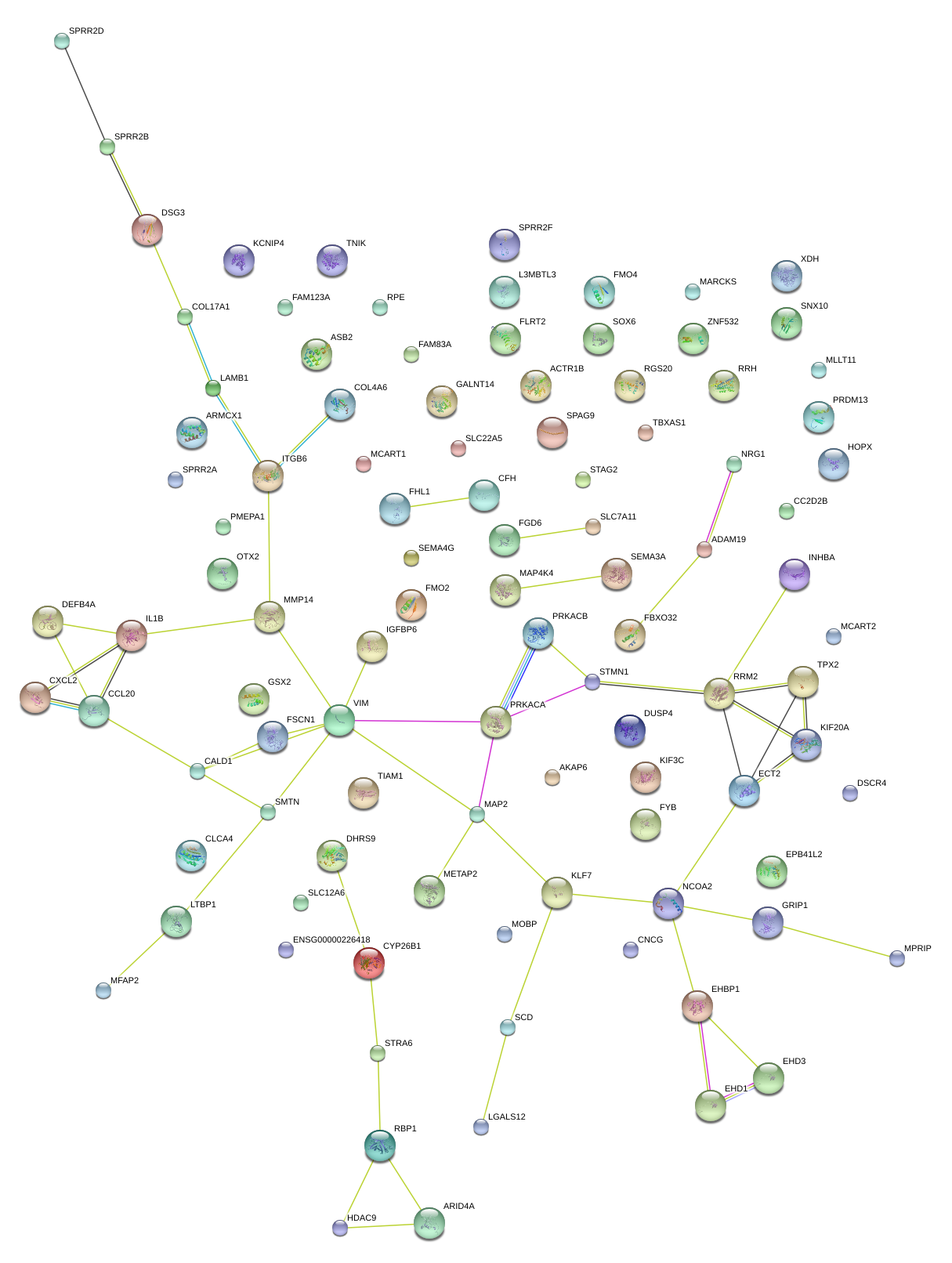
|
chr1_-_153029987
|
12.999
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr1_-_153044083
|
9.089
|
NM_001017418
|
SPRR2B
|
small proline-rich protein 2B
|
|
chr1_+_171154387
|
7.786
|
NM_001460
|
FMO2
|
flavin containing monooxygenase 2 (non-functional)
|
|
chr1_+_84647342
|
7.166
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr2_-_161056762
|
6.283
|
|
ITGB6
|
integrin, beta 6
|
|
chr1_-_153085963
|
5.128
|
NM_001014450
|
SPRR2A
SPRR2F
|
small proline-rich protein 2A
small proline-rich protein 2F
|
|
chr2_+_102508333
|
4.996
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr2_-_26205437
|
4.924
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr7_+_26400503
|
4.872
|
NM_001199837
|
SNX10
|
sorting nexin 10
|
|
chr7_-_41742666
|
4.810
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr1_-_153013593
|
4.744
|
NM_006945
|
SPRR2D
|
small proline-rich protein 2D
|
|
chr7_+_134464417
|
4.734
|
|
CALD1
|
caldesmon 1
|
|
chr7_-_83824216
|
4.357
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr2_-_26205327
|
4.345
|
|
KIF3C
|
kinesin family member 3C
|
|
chr8_-_124553253
|
3.904
|
|
FBXO32
|
F-box protein 32
|
|
chr2_+_102314162
|
3.893
|
NM_001242560
NM_004834
NM_145686
NM_001242559
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr12_-_67072881
|
3.889
|
NM_001178074
NM_021150
|
GRIP1
|
glutamate receptor interacting protein 1
|
|
chr6_+_114178516
|
3.778
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr8_+_32504250
|
3.651
|
NM_013959
|
NRG1
|
neuregulin 1
|
|
chr7_+_134464372
|
3.534
|
|
CALD1
|
caldesmon 1
|
|
chr11_-_16424391
|
3.516
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr8_+_124194751
|
3.515
|
NM_032899
NM_207006
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr15_-_34610928
|
3.509
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr7_-_107643330
|
3.489
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr2_+_102508377
|
3.472
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chrX_+_100805415
|
3.409
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr14_+_85996454
|
3.405
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr2_-_72374919
|
3.377
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr6_-_30655671
|
3.363
|
NM_133471
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr8_-_7274353
|
3.259
|
NM_001205266
|
DEFB4B
|
defensin, beta 4B
|
|
chr14_-_57272344
|
3.222
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr2_+_210444364
|
3.211
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr1_+_151032880
|
3.203
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr12_+_53491417
|
3.195
|
NM_002178
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr1_-_26233367
|
3.184
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr15_-_74494780
|
3.169
|
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr22_+_31477231
|
3.167
|
NM_006932
NM_134269
NM_134270
|
SMTN
|
smoothelin
|
|
chr1_+_151032866
|
3.161
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr2_-_161056573
|
3.157
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr6_+_100054649
|
3.129
|
NM_021620
|
PRDM13
|
PR domain containing 13
|
|
chr15_-_74495093
|
3.124
|
NM_001199042
NM_022369
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr6_+_130339715
|
3.119
|
NM_001007102
NM_032438
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr10_+_102732285
|
3.096
|
NM_001203244
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr5_+_137514416
|
3.086
|
NM_005733
|
KIF20A
|
kinesin family member 20A
|
|
chr8_+_54764367
|
3.014
|
NM_170587
|
RGS20
|
regulator of G-protein signaling 20
|
|
chr15_-_74494851
|
3.006
|
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr8_+_7752198
|
2.986
|
NM_004942
|
DEFB4A
|
defensin, beta 4A
|
|
chr1_+_151032683
|
2.976
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr5_+_137514753
|
2.964
|
|
KIF20A
|
kinesin family member 20A
|
|
chr2_-_31352489
|
2.921
|
NM_001253827
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chr7_+_5632435
|
2.909
|
NM_003088
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr12_+_53491448
|
2.907
|
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr2_+_33359607
|
2.900
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chrX_+_135251454
|
2.866
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr12_-_95611023
|
2.865
|
NM_018351
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chr7_+_5632453
|
2.817
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chrX_+_135251795
|
2.802
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr18_+_29027731
|
2.801
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr4_+_110749149
|
2.791
|
NM_006583
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog
|
|
chr17_-_49124128
|
2.777
|
NM_001251971
|
SPAG9
|
sperm associated antigen 9
|
|
chr1_+_87012758
|
2.777
|
NM_012128
|
CLCA4
|
chloride channel accessory 4
|
|
chr8_-_29206321
|
2.708
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr17_+_16945766
|
2.704
|
NM_015134
NM_201274
|
MPRIP
|
myosin phosphatase Rho interacting protein
|
|
chr2_+_169923531
|
2.670
|
NM_199204
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr10_+_105005643
|
2.646
|
NM_001143909
|
LOC729020
|
rcRPE
|
|
chr21_-_32931289
|
2.643
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr2_-_208031570
|
2.569
|
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr5_-_39203061
|
2.561
|
|
FYB
|
FYN binding protein
|
|
chr10_-_105845619
|
2.527
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr10_+_97759847
|
2.480
|
NM_001001732
NM_001159747
|
CC2D2B
|
coiled-coil and C2 domain containing 2B
|
|
chr3_-_139258373
|
2.410
|
|
RBP1
|
retinol binding protein 1, cellular
|
|
chr3_-_139258477
|
2.397
|
|
RBP1
|
retinol binding protein 1, cellular
|
|
chr2_-_113594325
|
2.378
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr4_-_47983430
|
2.371
|
NM_001142564
|
CNGA1
|
cyclic nucleotide gated channel alpha 1
|
|
chr1_-_17308072
|
2.370
|
NM_001135247
NM_017459
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr10_+_17271302
|
2.348
|
|
VIM
|
vimentin
|
|
chr8_+_124194873
|
2.340
|
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr10_-_105845556
|
2.312
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr2_-_31637598
|
2.262
|
NM_000379
|
XDH
|
xanthine dehydrogenase
|
|
chr10_+_17271276
|
2.258
|
|
VIM
|
vimentin
|
|
chr2_+_10262694
|
2.234
|
NM_001165931
|
RRM2
|
ribonucleotide reductase M2
|
|
chr18_-_29340698
|
2.232
|
NM_001034172
|
MCART2
|
mitochondrial carrier triple repeat 2
|
|
chr10_+_17271277
|
2.215
|
|
VIM
|
vimentin
|
|
chr4_+_54966197
|
2.194
|
NM_133267
|
GSX2
|
GS homeobox 2
|
|
chr14_+_23305816
|
2.175
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr20_+_30326903
|
2.140
|
NM_012112
|
TPX2
|
TPX2, microtubule-associated, homolog (Xenopus laevis)
|
|
chr10_+_17270469
|
2.109
|
|
VIM
|
vimentin
|
|
chr6_-_131291480
|
2.094
|
NM_001199389
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr4_-_139163222
|
2.089
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr12_-_122240827
|
2.067
|
|
|
|
|
chr5_-_157002739
|
2.031
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr3_+_172472246
|
2.028
|
NM_018098
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr4_-_57547845
|
2.027
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chrX_-_107682600
|
2.023
|
NM_001847
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chr2_+_10262853
|
2.000
|
NM_001034
|
RRM2
|
ribonucleotide reductase M2
|
|
chr14_+_23305792
|
1.993
|
NM_004995
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr10_+_102106771
|
1.988
|
NM_005063
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr2_+_62934030
|
1.970
|
NM_001142614
|
EHBP1
|
EH domain binding protein 1
|
|
chr18_+_56530677
|
1.958
|
|
ZNF532
|
zinc finger protein 532
|
|
chr3_+_39509069
|
1.955
|
NM_182935
|
MOBP
|
myelin-associated oligodendrocyte basic protein
|
|
chr4_-_20985547
|
1.954
|
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr2_+_228678556
|
1.942
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr14_+_32798478
|
1.939
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chrX_+_123234462
|
1.936
|
|
STAG2
|
stromal antigen 2
|
|
chr14_-_94423766
|
1.925
|
NM_016150
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr11_-_64647136
|
1.924
|
|
EHD1
|
EH-domain containing 1
|
|
chr20_-_56284945
|
1.920
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr7_+_18535351
|
1.918
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr13_-_25745620
|
1.918
|
|
FAM123A
|
family with sequence similarity 123A
|
|
chrX_+_135229536
|
1.913
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr7_+_18548899
|
1.903
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr7_+_139528951
|
1.900
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr14_-_65409445
|
1.872
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr7_-_148581297
|
1.857
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chrX_-_153599581
|
1.845
|
|
FLNA
|
filamin A, alpha
|
|
chr10_+_54074037
|
1.843
|
NM_012242
|
DKK1
|
dickkopf 1 homolog (Xenopus laevis)
|
|
chr1_-_6479880
|
1.842
|
NM_019089
|
HES2
|
hairy and enhancer of split 2 (Drosophila)
|
|
chr15_-_70390226
|
1.842
|
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila)
|
|
chr7_+_106505696
|
1.827
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr2_-_165424993
|
1.824
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr11_+_125496123
|
1.821
|
NM_001274
|
CHEK1
|
checkpoint kinase 1
|
|
chr3_+_19189969
|
1.821
|
NM_144633
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8
|
|
chr11_-_61584228
|
1.812
|
|
FADS1
|
fatty acid desaturase 1
|
|
chr3_-_123339178
|
1.808
|
|
MYLK
|
myosin light chain kinase
|
|
chr16_-_75529278
|
1.805
|
|
|
|
|
chr5_+_126112847
|
1.795
|
|
LMNB1
|
lamin B1
|
|
chr2_-_2334887
|
1.781
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr2_+_208576446
|
1.773
|
|
CCNYL1
|
cyclin Y-like 1
|
|
chr5_+_31193761
|
1.762
|
NM_004932
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr9_+_27109138
|
1.759
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr14_-_73493744
|
1.757
|
NM_021260
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr2_+_75061234
|
1.750
|
|
HK2
|
hexokinase 2
|
|
chr1_+_90098643
|
1.749
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr12_-_28124915
|
1.748
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr3_-_149095458
|
1.746
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr4_-_69111333
|
1.740
|
NM_182502
|
TMPRSS11B
|
transmembrane protease, serine 11B
|
|
chr9_-_72287097
|
1.734
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr11_+_129991582
|
1.734
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr5_-_16936290
|
1.732
|
|
MYO10
|
myosin X
|
|
chr7_+_28452143
|
1.720
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr13_+_111137249
|
1.711
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr1_-_36097137
|
1.708
|
NM_001199780
|
PSMB2
|
proteasome (prosome, macropain) subunit, beta type, 2
|
|
chr7_+_80267782
|
1.707
|
NM_000072
NM_001001548
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr3_-_123339423
|
1.703
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr12_-_8815479
|
1.697
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr5_+_126112819
|
1.694
|
NM_001198557
|
LMNB1
|
lamin B1
|
|
chr19_-_49865638
|
1.692
|
NM_003598
|
TEAD2
|
TEA domain family member 2
|
|
chr1_-_155177685
|
1.663
|
NM_001252607
NM_001252608
NM_007112
|
THBS3
|
thrombospondin 3
|
|
chr2_-_190044330
|
1.655
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr15_-_56209305
|
1.646
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr12_+_48876285
|
1.643
|
NM_152319
|
C12orf54
|
chromosome 12 open reading frame 54
|
|
chr4_-_57522469
|
1.643
|
NM_001145459
NM_139211
|
HOPX
|
HOP homeobox
|
|
chr1_+_84630575
|
1.642
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr3_+_189507448
|
1.637
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chr6_+_12290528
|
1.633
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr21_-_32201926
|
1.629
|
NM_181606
|
KRTAP7-1
|
keratin associated protein 7-1 (gene/pseudogene)
|
|
chr10_-_29754478
|
1.619
|
|
SVIL
|
supervillin
|
|
chr21_+_35190586
|
1.612
|
|
ITSN1
|
intersectin 1 (SH3 domain protein)
|
|
chr5_+_149865310
|
1.589
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr7_-_148581359
|
1.586
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr2_-_166060552
|
1.568
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr7_-_143059144
|
1.559
|
NM_014690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr1_-_153067000
|
1.558
|
NM_001024209
|
SPRR2E
|
small proline-rich protein 2E
|
|
chr7_+_94297448
|
1.558
|
|
PEG10
|
paternally expressed 10
|
|
chr6_-_64029847
|
1.555
|
NM_001143940
NM_016571
|
LGSN
|
lengsin, lens protein with glutamine synthetase domain
|
|
chr7_-_14030864
|
1.552
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chr16_+_55600583
|
1.550
|
NM_032330
|
CAPNS2
|
calpain, small subunit 2
|
|
chr12_+_93963597
|
1.547
|
NM_003877
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr3_+_189349215
|
1.539
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|
|
chr8_-_124553448
|
1.539
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr7_-_148581377
|
1.536
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr13_+_78109808
|
1.524
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr5_-_39424930
|
1.518
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr2_+_208576239
|
1.508
|
NM_001142300
NM_152523
|
CCNYL1
|
cyclin Y-like 1
|
|
chr4_+_156680125
|
1.506
|
NM_000857
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr20_-_5591487
|
1.503
|
NM_019593
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae)
|
|
chr15_+_67358194
|
1.500
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr11_-_71752099
|
1.486
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr16_+_58497548
|
1.473
|
NM_020465
|
NDRG4
|
NDRG family member 4
|
|
chr19_-_42931500
|
1.472
|
NM_005357
|
LIPE
|
lipase, hormone-sensitive
|
|
chr7_-_14031049
|
1.469
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr3_-_192635933
|
1.467
|
NM_178496
|
MB21D2
|
Mab-21 domain containing 2
|
|
chr21_+_30517897
|
1.463
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr7_+_111846642
|
1.460
|
NM_021994
|
ZNF277
|
zinc finger protein 277
|
|
chr16_-_90085786
|
1.443
|
NM_001042610
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1
|
|
chr9_+_116342939
|
1.442
|
NM_021106
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_-_95298251
|
1.436
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr17_-_7297826
|
1.425
|
|
PLSCR3
|
phospholipid scramblase 3
|
|
chr9_+_116343169
|
1.424
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr6_+_80714389
|
1.418
|
|
TTK
|
TTK protein kinase
|
|
chrX_+_41548225
|
1.409
|
NM_001097579
NM_005300
|
GPR34
|
G protein-coupled receptor 34
|
|
chr19_+_41257086
|
1.407
|
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A
|
|
chr5_-_114515733
|
1.405
|
NM_001017397
NM_001017398
NM_018700
|
TRIM36
|
tripartite motif containing 36
|
|
chr13_-_25745778
|
1.401
|
NM_152704
NM_199138
|
FAM123A
|
family with sequence similarity 123A
|
|
chr5_-_132161863
|
1.392
|
NM_133456
|
SHROOM1
|
shroom family member 1
|
|
chr12_+_49212253
|
1.388
|
NM_000725
NM_001206915
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr16_+_58497628
|
1.387
|
|
NDRG4
|
NDRG family member 4
|
|
chr17_-_10452939
|
1.386
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr3_-_119582422
|
1.384
|
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr15_-_70390229
|
1.381
|
NM_001105192
NM_005078
NM_020908
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila)
|