|
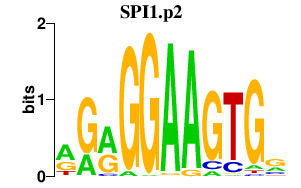
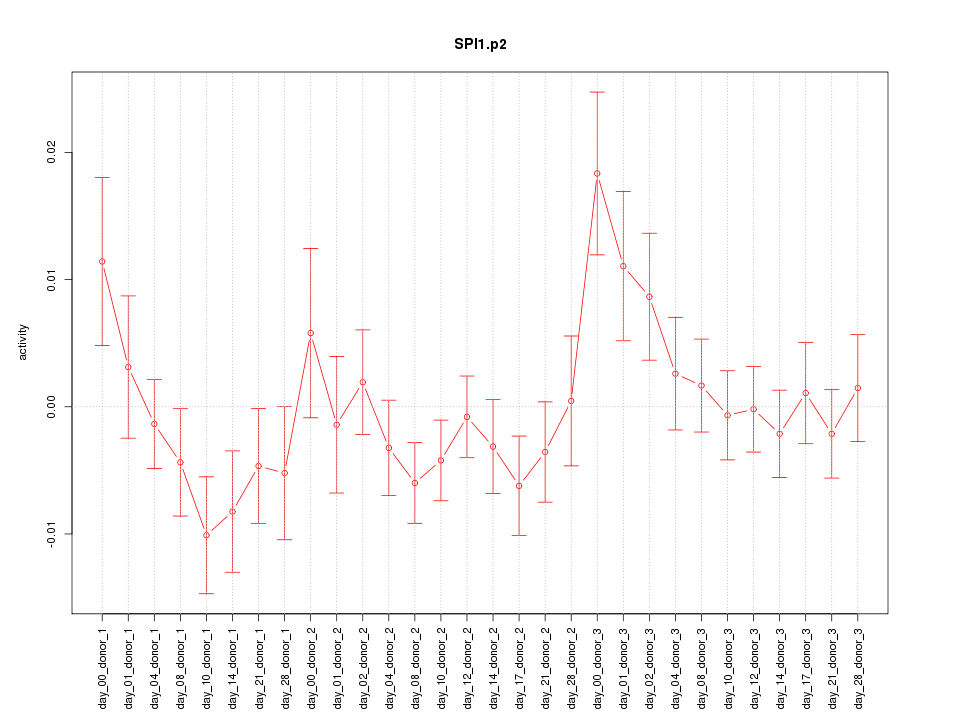
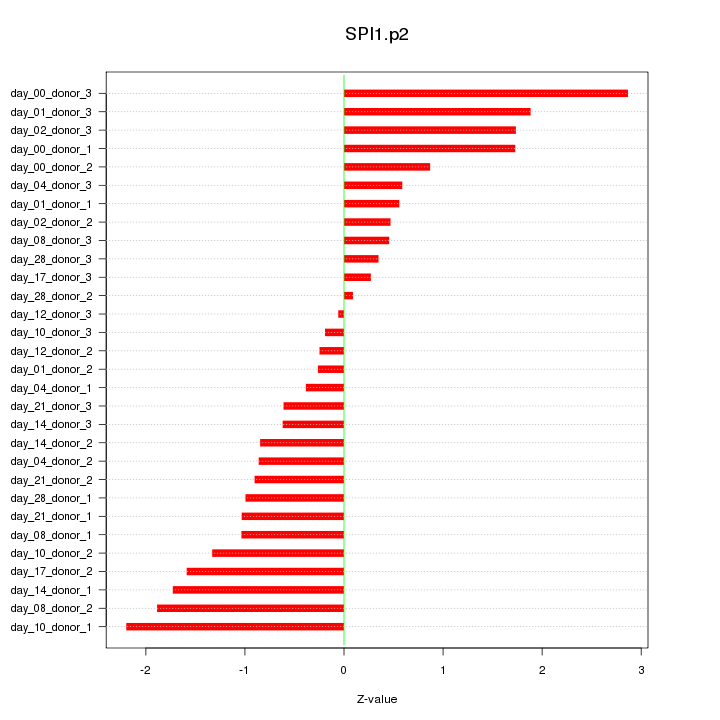
chr19_-_51504799
|
4.574
|
NM_007196
NM_144505
NM_144506
NM_144507
|
KLK8
|
kallikrein-related peptidase 8
|
|
chr19_-_36019194
|
3.583
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chr19_-_51472003
|
3.385
|
NM_001012964
NM_001012965
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr19_+_35645614
|
3.186
|
NM_014164
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr12_-_25101993
|
3.044
|
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr6_-_30652434
|
3.034
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr19_+_35645640
|
3.021
|
NM_144779
NM_001164605
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr6_-_30654978
|
2.956
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr9_-_107690526
|
2.685
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr6_-_30654406
|
2.647
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr20_+_3776905
|
2.616
|
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr19_-_51456159
|
2.608
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr15_+_63335086
|
2.573
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr1_+_150480584
|
2.528
|
|
ECM1
|
extracellular matrix protein 1
|
|
chr15_+_67358161
|
2.528
|
|
SMAD3
|
SMAD family member 3
|
|
chr12_+_41086189
|
2.491
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr12_-_25101923
|
2.474
|
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr5_-_39219578
|
2.378
|
NM_001465
NM_199335
|
FYB
|
FYN binding protein
|
|
chr4_+_8200918
|
2.373
|
NM_018986
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1
|
|
chr19_-_51472816
|
2.281
|
NM_002774
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr7_-_100860991
|
2.206
|
NM_001084
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr7_+_27135672
|
2.201
|
|
HOTAIRM1
|
HOXA transcript antisense RNA, myeloid-specific 1 (non-protein coding)
|
|
chr2_+_31456879
|
2.197
|
NM_014600
|
EHD3
|
EH-domain containing 3
|
|
chr17_-_45933138
|
2.188
|
NM_199262
|
SP6
|
Sp6 transcription factor
|
|
chr18_+_56338818
|
2.186
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr17_+_1959512
|
2.142
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr10_-_5541401
|
2.118
|
NM_017422
|
CALML5
|
calmodulin-like 5
|
|
chr17_+_65373574
|
2.108
|
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1
|
|
chr19_+_35646006
|
1.908
|
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr7_-_100860865
|
1.905
|
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr2_-_72374919
|
1.861
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr7_-_100860841
|
1.842
|
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr12_+_107712151
|
1.794
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr5_-_172755195
|
1.789
|
|
STC2
|
stanniocalcin 2
|
|
chr12_+_53443834
|
1.764
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr11_+_118827007
|
1.754
|
NM_006760
|
UPK2
|
uroplakin 2
|
|
chrX_-_15872905
|
1.734
|
|
|
|
|
chr2_-_65659634
|
1.690
|
NM_181784
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr18_+_21452981
|
1.644
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr10_-_135090325
|
1.627
|
NM_001109
NM_001164489
NM_001164490
|
ADAM8
|
ADAM metallopeptidase domain 8
|
|
chr6_+_86159301
|
1.623
|
NM_001204813
NM_002526
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr20_-_23030141
|
1.613
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr1_+_230202966
|
1.604
|
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2)
|
|
chr11_-_66725790
|
1.588
|
NM_000920
NM_001040716
|
PC
|
pyruvate carboxylase
|
|
chr18_+_56338783
|
1.540
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr1_-_159923961
|
1.516
|
NM_001146172
NM_001146173
NM_033438
|
SLAMF9
|
SLAM family member 9
|
|
chr2_-_238322840
|
1.497
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_65659115
|
1.484
|
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr6_+_43738757
|
1.475
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr6_-_30655078
|
1.471
|
NM_001134870
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr19_-_8642232
|
1.452
|
|
MYO1F
|
myosin IF
|
|
chr7_+_18126543
|
1.440
|
NM_001204144
|
HDAC9
|
histone deacetylase 9
|
|
chr2_-_113542970
|
1.439
|
NM_000575
|
IL1A
|
interleukin 1, alpha
|
|
chr7_-_130353561
|
1.439
|
|
COPG2
|
coatomer protein complex, subunit gamma 2
|
|
chr7_-_130353579
|
1.436
|
NM_012133
|
COPG2
|
coatomer protein complex, subunit gamma 2
|
|
chr1_+_17531619
|
1.424
|
NM_013358
|
PADI1
|
peptidyl arginine deiminase, type I
|
|
chr2_-_238322770
|
1.415
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_238322815
|
1.393
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chrX_+_135278912
|
1.390
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chrX_-_47489243
|
1.387
|
NM_001145252
|
CFP
|
complement factor properdin
|
|
chr18_+_56338678
|
1.387
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr11_+_61595633
|
1.380
|
NM_004265
|
FADS2
|
fatty acid desaturase 2
|
|
chr17_-_41623652
|
1.377
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr9_+_34653911
|
1.372
|
NM_004512
NM_147162
|
IL11RA
|
interleukin 11 receptor, alpha
|
|
chrX_-_15872921
|
1.366
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr12_-_123466195
|
1.361
|
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|
|
chr1_-_197115566
|
1.352
|
NM_001206846
NM_018136
|
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila)
|
|
chr2_+_74120092
|
1.345
|
NM_001199893
NM_001615
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr12_-_54785018
|
1.336
|
NM_001130967
NM_001130968
|
ZNF385A
|
zinc finger protein 385A
|
|
chr12_+_96588000
|
1.331
|
NM_005230
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2)
|
|
chr17_+_7210929
|
1.331
|
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr5_-_39270708
|
1.325
|
NM_001243093
|
FYB
|
FYN binding protein
|
|
chr4_-_111119752
|
1.308
|
NM_001130721
NM_024090
|
ELOVL6
|
ELOVL fatty acid elongase 6
|
|
chr1_+_32050510
|
1.288
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr7_-_107643330
|
1.285
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr15_+_67358194
|
1.284
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr22_+_31489343
|
1.283
|
|
SMTN
|
smoothelin
|
|
chr8_+_22022644
|
1.276
|
NM_001199
NM_006129
|
BMP1
|
bone morphogenetic protein 1
|
|
chr11_-_58343308
|
1.271
|
NM_004811
|
LPXN
|
leupaxin
|
|
chr2_-_31361284
|
1.270
|
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chr7_-_100860719
|
1.263
|
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr11_-_64647136
|
1.262
|
|
EHD1
|
EH-domain containing 1
|
|
chr2_+_74120132
|
1.255
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr9_+_128509616
|
1.240
|
NM_006195
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr12_+_96588254
|
1.231
|
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2)
|
|
chr2_-_37193609
|
1.225
|
NM_003162
|
STRN
|
striatin, calmodulin binding protein
|
|
chr19_+_48828820
|
1.215
|
|
EMP3
|
epithelial membrane protein 3
|
|
chr8_+_22022677
|
1.214
|
|
BMP1
|
bone morphogenetic protein 1
|
|
chr17_-_41623246
|
1.198
|
NM_001986
|
ETV4
|
ets variant 4
|
|
chr7_+_106505696
|
1.185
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr1_-_6479880
|
1.168
|
NM_019089
|
HES2
|
hairy and enhancer of split 2 (Drosophila)
|
|
chr11_-_2160199
|
1.158
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr16_+_23690190
|
1.157
|
|
PLK1
|
polo-like kinase 1
|
|
chr11_-_112034671
|
1.140
|
NM_001243211
NM_001562
|
IL18
|
interleukin 18 (interferon-gamma-inducing factor)
|
|
chr6_+_74405764
|
1.137
|
|
CD109
|
CD109 molecule
|
|
chr6_-_32160637
|
1.135
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr1_-_20812271
|
1.133
|
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr7_-_100860615
|
1.132
|
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr1_+_8384389
|
1.127
|
NM_001080397
|
SLC45A1
|
solute carrier family 45, member 1
|
|
chr1_+_154300275
|
1.118
|
NM_020452
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr11_+_61595772
|
1.111
|
|
FADS2
|
fatty acid desaturase 2
|
|
chr5_-_127873734
|
1.105
|
NM_001999
|
FBN2
|
fibrillin 2
|
|
chr20_+_361272
|
1.096
|
NM_021158
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr1_-_231175971
|
1.094
|
NM_198552
|
FAM89A
|
family with sequence similarity 89, member A
|
|
chr7_-_130353490
|
1.087
|
|
COPG2
|
coatomer protein complex, subunit gamma 2
|
|
chr1_-_155948934
|
1.080
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr20_+_33759773
|
1.076
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr16_+_23690191
|
1.076
|
NM_005030
|
PLK1
|
polo-like kinase 1
|
|
chr17_-_36413177
|
1.067
|
|
LOC440434
|
aminopeptidase puromycin sensitive pseudogene
|
|
chr22_-_37640187
|
1.067
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr4_+_79472851
|
1.057
|
|
ANXA3
|
annexin A3
|
|
chr11_+_2920910
|
1.057
|
NM_183233
|
SLC22A18
|
solute carrier family 22, member 18
|
|
chr6_+_86159738
|
1.045
|
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr9_-_101017492
|
1.043
|
|
TBC1D2
|
TBC1 domain family, member 2
|
|
chr11_+_1861431
|
1.042
|
NM_001145841
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr12_-_48152860
|
1.039
|
NM_001098531
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr6_+_37137882
|
1.034
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr7_+_100797685
|
1.033
|
NM_001283
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit
|
|
chr8_-_10587942
|
1.027
|
NM_031439
|
SOX7
|
SRY (sex determining region Y)-box 7
|
|
chr20_+_3776385
|
1.027
|
NM_004358
NM_021872
NM_021873
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr12_-_49365545
|
1.023
|
NM_003394
|
WNT10B
|
wingless-type MMTV integration site family, member 10B
|
|
chr9_+_74526369
|
1.021
|
NM_182505
|
C9orf85
|
chromosome 9 open reading frame 85
|
|
chr12_-_109915127
|
1.013
|
NM_031954
|
KCTD10
|
potassium channel tetramerisation domain containing 10
|
|
chr19_+_48828627
|
1.011
|
NM_001425
|
EMP3
|
epithelial membrane protein 3
|
|
chrX_-_47489703
|
1.008
|
NM_002621
|
CFP
|
complement factor properdin
|
|
chr12_-_48152180
|
0.985
|
NM_001098532
NM_006105
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr14_+_55034633
|
0.982
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr15_+_63334753
|
0.979
|
NM_000366
NM_001018004
NM_001018005
NM_001018006
NM_001018007
NM_001018020
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chrX_-_119694492
|
0.979
|
|
CUL4B
|
cullin 4B
|
|
chr11_+_129245834
|
0.979
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr2_-_70780621
|
0.974
|
|
TGFA
|
transforming growth factor, alpha
|
|
chr12_-_72057676
|
0.972
|
NM_144982
|
ZFC3H1
|
zinc finger, C3H1-type containing
|
|
chr5_+_86563594
|
0.968
|
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr5_-_140998480
|
0.966
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr19_-_36004553
|
0.960
|
NM_001126056
NM_001126057
NM_001126058
NM_001190347
NM_001190348
NM_001190349
NM_033317
|
DMKN
|
dermokine
|
|
chr17_+_2699731
|
0.958
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr15_+_90744506
|
0.956
|
NM_198925
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr2_-_30144431
|
0.953
|
NM_004304
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase
|
|
chr4_+_79472741
|
0.950
|
NM_005139
|
ANXA3
|
annexin A3
|
|
chr15_+_93443559
|
0.948
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr17_-_7155258
|
0.943
|
NM_015343
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1
|
|
chr8_+_25316736
|
0.937
|
|
CDCA2
|
cell division cycle associated 2
|
|
chr1_-_159915382
|
0.931
|
NM_001135050
NM_020789
|
IGSF9
|
immunoglobulin superfamily, member 9
|
|
chr6_-_75994496
|
0.930
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr16_+_23690160
|
0.929
|
|
PLK1
|
polo-like kinase 1
|
|
chr9_+_34990715
|
0.920
|
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chrX_+_99899388
|
0.920
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr1_-_155959860
|
0.918
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr17_-_26903854
|
0.917
|
|
ALDOC
|
aldolase C, fructose-bisphosphate
|
|
chr16_+_14726828
|
0.916
|
|
BFAR
|
bifunctional apoptosis regulator
|
|
chr17_-_39768939
|
0.915
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr12_-_120687956
|
0.912
|
NM_025157
|
PXN
|
paxillin
|
|
chr17_+_65373362
|
0.911
|
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1
|
|
chr20_-_44540685
|
0.905
|
|
PLTP
|
phospholipid transfer protein
|
|
chr9_-_107690336
|
0.904
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr17_+_57232859
|
0.899
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr1_-_168106535
|
0.898
|
|
GPR161
|
G protein-coupled receptor 161
|
|
chr15_+_38544119
|
0.893
|
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr2_-_228029274
|
0.890
|
NM_000092
|
COL4A4
|
collagen, type IV, alpha 4
|
|
chr3_+_50654589
|
0.890
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr16_-_74734687
|
0.889
|
NM_001142497
NM_152649
|
MLKL
|
mixed lineage kinase domain-like
|
|
chr16_+_83986826
|
0.889
|
NM_182981
|
OSGIN1
|
oxidative stress induced growth inhibitor 1
|
|
chr9_-_114937464
|
0.881
|
NM_022486
|
SUSD1
|
sushi domain containing 1
|
|
chr1_+_155178489
|
0.879
|
NM_002455
NM_198883
|
MTX1
|
metaxin 1
|
|
chr10_+_75673346
|
0.874
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr16_+_30194730
|
0.873
|
NM_001193333
NM_007074
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr7_-_23509972
|
0.872
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr10_-_33623978
|
0.868
|
|
NRP1
|
neuropilin 1
|
|
chr12_-_53600999
|
0.867
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr5_-_172755418
|
0.865
|
|
STC2
|
stanniocalcin 2
|
|
chr16_+_23690234
|
0.865
|
|
PLK1
|
polo-like kinase 1
|
|
chrX_-_40594663
|
0.862
|
|
MED14
|
mediator complex subunit 14
|
|
chr11_+_72980938
|
0.860
|
NM_176798
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6
|
|
chr12_+_1800146
|
0.856
|
|
ADIPOR2
|
adiponectin receptor 2
|
|
chr6_+_43543877
|
0.856
|
NM_006502
|
POLH
|
polymerase (DNA directed), eta
|
|
chr5_+_135364583
|
0.843
|
NM_000358
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr22_+_38453478
|
0.841
|
|
PICK1
|
protein interacting with PRKCA 1
|
|
chr5_-_140013034
|
0.841
|
NM_000591
|
CD14
|
CD14 molecule
|
|
chr3_-_111851967
|
0.838
|
NM_001190259
NM_001190260
NM_152785
|
GCET2
|
germinal center expressed transcript 2
|
|
chr17_+_25621105
|
0.835
|
NM_015626
NM_134265
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr5_-_72744351
|
0.832
|
NM_004472
|
FOXD1
|
forkhead box D1
|
|
chr4_-_143767603
|
0.827
|
NM_001101669
NM_003866
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa
|
|
chr6_-_43595079
|
0.823
|
|
GTPBP2
|
GTP binding protein 2
|
|
chrX_+_48367346
|
0.822
|
NM_022825
NM_203473
|
PORCN
|
porcupine homolog (Drosophila)
|
|
chr6_-_112575880
|
0.820
|
|
LAMA4
|
laminin, alpha 4
|
|
chr14_+_68086633
|
0.816
|
|
ARG2
|
arginase, type II
|
|
chr5_+_92918924
|
0.813
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr12_+_49716970
|
0.812
|
NM_001100620
NM_005480
|
TROAP
|
trophinin associated protein (tastin)
|
|
chr11_+_130184326
|
0.810
|
|
FLJ34521
|
uncharacterized LOC646383
|
|
chr1_-_209957872
|
0.808
|
NM_152485
|
C1orf74
|
chromosome 1 open reading frame 74
|
|
chr17_-_74497406
|
0.804
|
NM_024599
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chr3_-_48130633
|
0.804
|
NM_001134364
NM_002375
NM_030885
|
MAP4
|
microtubule-associated protein 4
|
|
chr11_-_31832784
|
0.801
|
|
PAX6
|
paired box 6
|
|
chr17_+_1958354
|
0.796
|
NM_006497
|
HIC1
|
hypermethylated in cancer 1
|
|
chr19_+_16187047
|
0.793
|
NM_003290
|
TPM4
|
tropomyosin 4
|
|
chr7_-_99698278
|
0.792
|
NM_182776
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr7_-_98030165
|
0.789
|
|
BAIAP2L1
|
BAI1-associated protein 2-like 1
|
|
chr1_-_153363544
|
0.789
|
NM_002964
|
S100A8
|
S100 calcium binding protein A8
|
|
chr17_-_62657997
|
0.788
|
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr5_+_86564044
|
0.787
|
NM_002890
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|