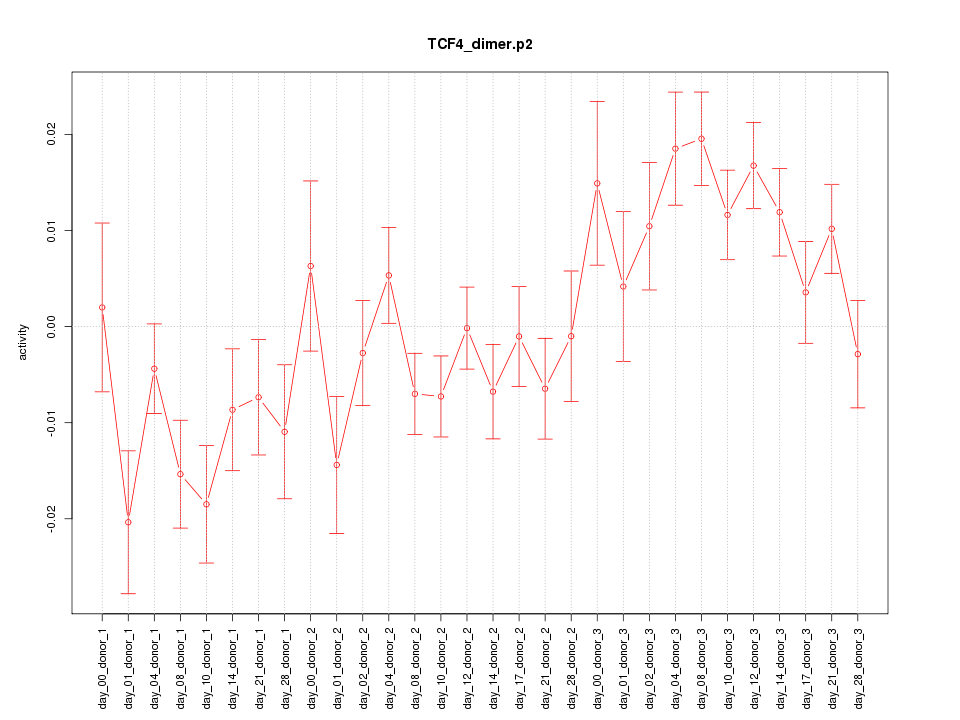
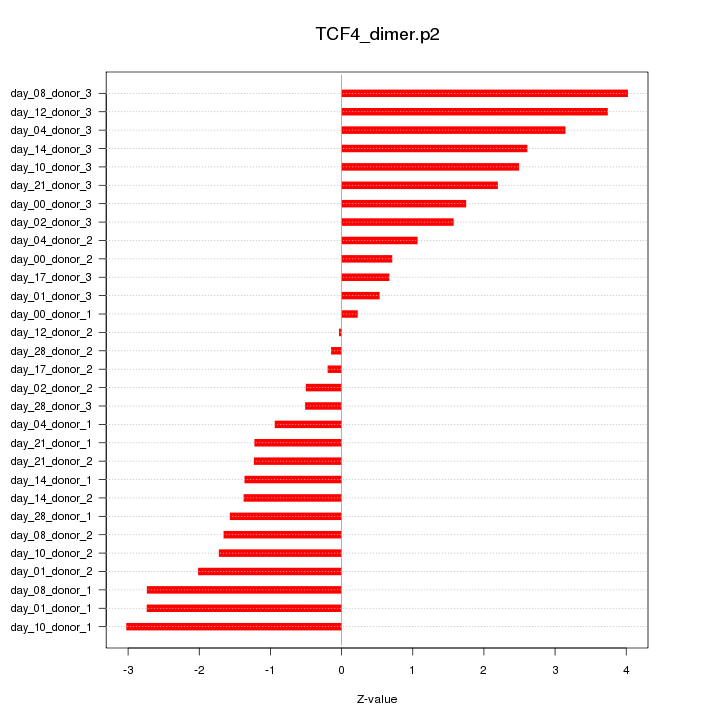
|
chr12_-_91505215
|
6.871
|
|
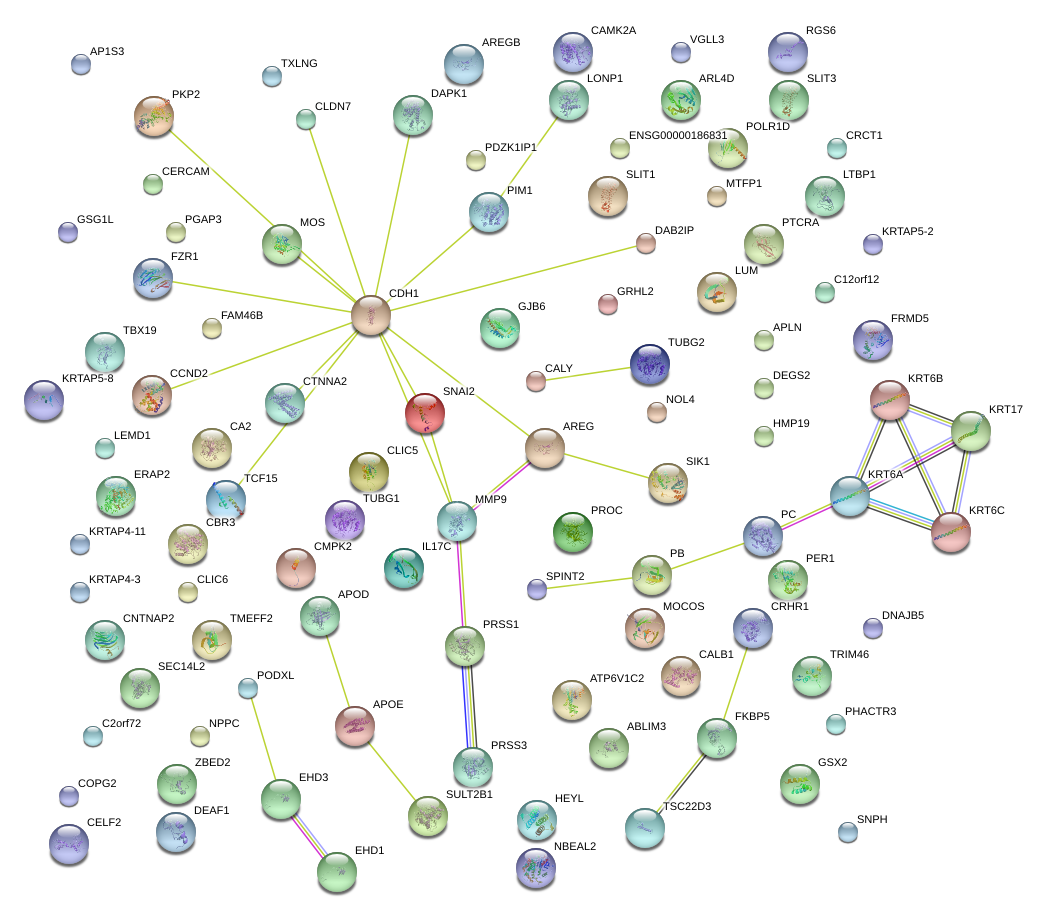
LUM
|
lumican
|
|
chr12_-_91505541
|
6.670
|
NM_002345
|
LUM
|
lumican
|
|
chr17_-_39274587
|
4.166
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr10_-_135150368
|
4.158
|
|
CALY
|
calcyon neuron-specific vesicular protein
|
|
chr10_-_135150474
|
3.963
|
NM_015722
|
CALY
|
calcyon neuron-specific vesicular protein
|
|
chr5_+_148520985
|
3.270
|
NM_014945
|
ABLIM3
|
actin binding LIM protein family, member 3
|
|
chr8_+_102504661
|
3.175
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr8_+_102504836
|
2.955
|
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr5_+_173472692
|
2.722
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr1_-_27339326
|
2.545
|
NM_052943
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr21_+_37507209
|
2.491
|
NM_001236
|
CBR3
|
carbonyl reductase 3
|
|
chr2_+_33359607
|
2.343
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr4_+_75310852
|
2.320
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr17_-_39324423
|
2.202
|
NM_033187
|
KRTAP4-3
|
keratin associated protein 4-3
|
|
chr3_-_111314181
|
2.117
|
NM_024508
|
ZBED2
|
zinc finger, BED-type containing 2
|
|
chr20_-_590909
|
2.099
|
NM_004609
|
TCF15
|
transcription factor 15 (basic helix-loop-helix)
|
|
chr16_+_68771304
|
2.075
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chrX_+_16804531
|
2.068
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr1_+_182992694
|
2.066
|
|
|
|
|
chr6_-_45983279
|
2.065
|
NM_016929
|
CLIC5
|
chloride intracellular channel 5
|
|
chr12_+_4382882
|
2.019
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr4_+_54966197
|
2.008
|
NM_133267
|
GSX2
|
GS homeobox 2
|
|
chr2_+_79740059
|
1.964
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr1_-_27339448
|
1.962
|
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr11_-_66725790
|
1.927
|
NM_000920
NM_001040716
|
PC
|
pyruvate carboxylase
|
|
chr20_+_58179601
|
1.915
|
NM_080672
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr22_+_30792929
|
1.874
|
NM_001204204
NM_012429
NM_033382
|
SEC14L2
|
SEC14-like 2 (S. cerevisiae)
|
|
chr11_+_71249070
|
1.866
|
NM_021046
|
KRTAP5-8
|
keratin associated protein 5-8
|
|
chr1_-_47655603
|
1.839
|
|
PDZK1IP1
|
PDZK1 interacting protein 1
|
|
chr17_-_7165788
|
1.810
|
|
CLDN7
|
claudin 7
|
|
chr2_+_10861740
|
1.809
|
NM_001039362
NM_144583
|
ATP6V1C2
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C2
|
|
chr4_+_75480628
|
1.800
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr13_-_20806533
|
1.768
|
NM_001110219
NM_001110220
NM_001110221
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr2_-_7005764
|
1.766
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr11_-_64646053
|
1.755
|
NM_006795
|
EHD1
|
EH-domain containing 1
|
|
chr10_-_98945379
|
1.744
|
|
SLIT1
|
slit homolog 1 (Drosophila)
|
|
chrX_-_107018889
|
1.744
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr7_+_148036528
|
1.735
|
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr1_-_47655723
|
1.707
|
NM_005764
|
PDZK1IP1
|
PDZK1 interacting protein 1
|
|
chr9_+_90112754
|
1.688
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr11_-_64645925
|
1.676
|
|
EHD1
|
EH-domain containing 1
|
|
chr6_-_35696359
|
1.669
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr15_-_44487237
|
1.652
|
NM_032892
|
FRMD5
|
FERM domain containing 5
|
|
chr15_-_44487468
|
1.638
|
|
FRMD5
|
FERM domain containing 5
|
|
chr12_-_33049664
|
1.630
|
NM_001005242
NM_004572
|
PKP2
|
plakophilin 2
|
|
chr1_-_205391180
|
1.624
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr2_-_224702300
|
1.613
|
NM_001039569
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit
|
|
chr12_-_52845862
|
1.610
|
NM_005555
|
KRT6B
|
keratin 6B
|
|
chr9_+_131185129
|
1.603
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr9_+_34990715
|
1.592
|
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr1_+_155147051
|
1.589
|
|
TRIM46
|
tripartite motif containing 46
|
|
chr17_+_40811261
|
1.579
|
NM_016437
|
TUBG2
|
tubulin, gamma 2
|
|
chr12_-_91348950
|
1.560
|
NM_152638
|
C12orf12
|
chromosome 12 open reading frame 12
|
|
chr2_-_224702200
|
1.554
|
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit
|
|
chr14_-_100625931
|
1.535
|
NM_206918
|
DEGS2
|
delta(4)-desaturase, sphingolipid 2
|
|
chr3_-_45267620
|
1.526
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr8_+_86376130
|
1.526
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr3_-_87040246
|
1.526
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr8_-_57026540
|
1.501
|
NM_005372
|
MOS
|
v-mos Moloney murine sarcoma viral oncogene homolog
|
|
chr17_-_8059637
|
1.487
|
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr5_-_138725544
|
1.485
|
NM_016459
|
MZB1
|
marginal zone B and B1 cell-specific protein
|
|
chr20_+_1246959
|
1.479
|
NM_014723
|
SNPH
|
syntaphilin
|
|
chr3_-_45267068
|
1.453
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr21_-_44845920
|
1.451
|
|
SIK1
|
salt-inducible kinase 1
|
|
chr9_+_124505063
|
1.446
|
NM_138709
|
DAB2IP
|
DAB2 interacting protein
|
|
chr5_+_96212167
|
1.446
|
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2
|
|
chr3_-_195310985
|
1.442
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr8_-_91094924
|
1.442
|
NM_004929
|
CALB1
|
calbindin 1, 28kDa
|
|
chr9_+_33795558
|
1.435
|
NM_002771
|
PRSS3
|
protease, serine, 3
|
|
chr17_-_7166263
|
1.433
|
NM_001185023
NM_001307
|
CLDN7
|
claudin 7
|
|
chr19_+_49055421
|
1.413
|
NM_177973
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1
|
|
chr4_+_100871625
|
1.410
|
|
LOC256880
|
uncharacterized LOC256880
|
|
chr3_+_47021146
|
1.408
|
NM_015175
|
NBEAL2
|
neurobeachin-like 2
|
|
chr6_+_37137882
|
1.405
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr1_-_40098639
|
1.403
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr10_+_11206928
|
1.398
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr4_-_80994183
|
1.389
|
|
|
|
|
chr5_-_149669400
|
1.388
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chrX_-_128788913
|
1.381
|
NM_017413
|
APLN
|
apelin
|
|
chr7_-_130353490
|
1.369
|
|
COPG2
|
coatomer protein complex, subunit gamma 2
|
|
chr16_-_28074776
|
1.361
|
NM_001109763
|
GSG1L
|
GSG1-like
|
|
chr19_+_38755422
|
1.336
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr19_+_38755238
|
1.335
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr5_+_96211643
|
1.332
|
NM_001130140
NM_022350
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2
|
|
chr2_-_193059249
|
1.315
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr6_+_42883726
|
1.310
|
NM_001243168
NM_001243169
NM_001243170
NM_138296
|
PTCRA
|
pre T-cell antigen receptor alpha
|
|
chr2_+_231902194
|
1.304
|
NM_001144994
|
C2orf72
|
chromosome 2 open reading frame 72
|
|
chr18_-_31803514
|
1.302
|
NM_001198546
NM_001198548
NM_003787
|
NOL4
|
nucleolar protein 4
|
|
chr13_+_28194846
|
1.299
|
NM_001206559
|
POLR1D
|
polymerase (RNA) I polypeptide D, 16kDa
|
|
chr11_-_694842
|
1.295
|
|
DEAF1
|
deformed epidermal autoregulatory factor 1 (Drosophila)
|
|
chr10_+_11060069
|
1.291
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr10_-_98945580
|
1.287
|
|
SLIT1
|
slit homolog 1 (Drosophila)
|
|
chr16_+_68771214
|
1.283
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr10_+_11060105
|
1.276
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr16_+_88705000
|
1.276
|
NM_013278
|
IL17C
|
interleukin 17C
|
|
chr8_-_49833823
|
1.274
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr20_+_44637539
|
1.272
|
NM_004994
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase)
|
|
chr19_+_38755097
|
1.266
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr14_+_72398816
|
1.263
|
NM_001204423
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr10_+_11207053
|
1.248
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr2_-_232790958
|
1.244
|
NM_024409
|
NPPC
|
natriuretic peptide C
|
|
chrX_-_107019001
|
1.235
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_+_152486977
|
1.233
|
NM_019060
|
CRCT1
|
cysteine-rich C-terminal 1
|
|
chr1_-_144931707
|
1.233
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr17_+_70117154
|
1.229
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr9_+_34990266
|
1.224
|
NM_001135005
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr20_+_43029973
|
1.221
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr1_+_86046434
|
1.219
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr11_-_65667860
|
1.210
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr10_-_103347978
|
1.210
|
NM_001174084
NM_001174085
NM_013274
|
POLL
|
polymerase (DNA directed), lambda
|
|
chr12_+_41086189
|
1.207
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr14_-_51411118
|
1.201
|
NM_001163940
NM_002863
|
PYGL
|
phosphorylase, glycogen, liver
|
|
chr20_-_33734894
|
1.194
|
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2
|
|
chr6_-_112194430
|
1.193
|
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr5_-_149669187
|
1.186
|
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr4_-_41216455
|
1.180
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr1_+_168250277
|
1.178
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr3_+_30648372
|
1.176
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr9_-_131940481
|
1.176
|
|
IER5L
|
immediate early response 5-like
|
|
chr3_-_195310770
|
1.171
|
|
APOD
|
apolipoprotein D
|
|
chr10_-_103347882
|
1.169
|
|
POLL
|
polymerase (DNA directed), lambda
|
|
chr20_+_43029936
|
1.153
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr19_+_54926721
|
1.146
|
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chr17_+_28705960
|
1.145
|
|
CPD
|
carboxypeptidase D
|
|
chr9_-_22009270
|
1.140
|
NM_004936
NM_078487
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4)
|
|
chr20_+_43030005
|
1.140
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr19_+_54926604
|
1.128
|
NM_001005367
NM_001201461
NM_020659
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chr17_+_16945766
|
1.126
|
NM_015134
NM_201274
|
MPRIP
|
myosin phosphatase Rho interacting protein
|
|
chr19_-_45826131
|
1.124
|
|
CKM
|
creatine kinase, muscle
|
|
chr2_+_85980600
|
1.123
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr7_+_27135672
|
1.123
|
|
HOTAIRM1
|
HOXA transcript antisense RNA, myeloid-specific 1 (non-protein coding)
|
|
chr10_-_28591844
|
1.122
|
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7)
|
|
chr2_+_27505296
|
1.119
|
NM_032546
NM_187841
|
TRIM54
|
tripartite motif containing 54
|
|
chr7_+_80231503
|
1.112
|
NM_001001547
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr11_-_65667809
|
1.109
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr19_-_57183122
|
1.107
|
NM_001005850
|
ZNF835
|
zinc finger protein 835
|
|
chr19_+_48673936
|
1.107
|
|
C19orf68
|
chromosome 19 open reading frame 68
|
|
chr16_+_68679198
|
1.106
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr17_-_41174431
|
1.103
|
NM_006373
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr12_+_110205816
|
1.101
|
|
C12orf34
|
chromosome 12 open reading frame 34
|
|
chr20_-_33735060
|
1.096
|
NM_001145025
NM_018217
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2
|
|
chr15_+_63340314
|
1.091
|
NM_001018008
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr3_+_50712671
|
1.090
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr1_-_161520412
|
1.087
|
NM_001127593
NM_001127595
NM_001127596
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr22_-_36635999
|
1.086
|
NM_145637
|
APOL2
|
apolipoprotein L, 2
|
|
chr19_-_2308155
|
1.084
|
NM_001101391
|
LINGO3
|
leucine rich repeat and Ig domain containing 3
|
|
chr16_+_68679229
|
1.082
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr10_+_11059854
|
1.080
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr5_-_44388666
|
1.077
|
NM_004465
|
FGF10
|
fibroblast growth factor 10
|
|
chr8_-_145016691
|
1.076
|
NM_201383
|
PLEC
|
plectin
|
|
chr11_-_65667858
|
1.076
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr12_-_29936685
|
1.074
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chrX_+_1387692
|
1.073
|
NM_001161529
NM_001161531
NM_001161532
NM_006140
NM_172245
NM_172246
NM_172249
|
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
|
|
chr19_+_41107220
|
1.072
|
NM_001042545
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr16_+_11439279
|
1.070
|
NM_152308
|
RMI2
|
RMI2, RecQ mediated genome instability 2, homolog (S. cerevisiae)
|
|
chr14_-_107083398
|
1.069
|
|
IGH@
IGHA1
IGHG1
|
immunoglobulin heavy locus
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chrX_-_136113832
|
1.067
|
NM_054021
|
GPR101
|
G protein-coupled receptor 101
|
|
chr16_+_11439311
|
1.067
|
|
RMI2
|
RMI2, RecQ mediated genome instability 2, homolog (S. cerevisiae)
|
|
chr4_-_57522469
|
1.062
|
NM_001145459
NM_139211
|
HOPX
|
HOP homeobox
|
|
chr11_+_118401755
|
1.060
|
NM_001144034
NM_001144035
NM_001144036
NM_001144037
NM_001144038
NM_032780
|
TMEM25
|
transmembrane protein 25
|
|
chr2_+_223289285
|
1.060
|
NM_152386
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2
|
|
chr19_+_16187047
|
1.058
|
NM_003290
|
TPM4
|
tropomyosin 4
|
|
chr17_-_41174402
|
1.056
|
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr1_+_110210643
|
1.053
|
NM_000848
NM_001142368
|
GSTM2
|
glutathione S-transferase mu 2 (muscle)
|
|
chr17_-_80042414
|
1.050
|
|
FASN
|
fatty acid synthase
|
|
chr7_-_1595746
|
1.048
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr15_+_45722762
|
1.047
|
NM_032413
NM_197955
|
C15orf48
|
chromosome 15 open reading frame 48
|
|
chr9_-_104357202
|
1.044
|
NM_147180
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta
|
|
chr14_+_72399149
|
1.044
|
NM_001204424
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr6_+_47445440
|
1.042
|
NM_012120
|
CD2AP
|
CD2-associated protein
|
|
chr16_+_68771192
|
1.040
|
NM_004360
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr17_-_41174327
|
1.039
|
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr9_+_136399877
|
1.034
|
NM_014694
|
ADAMTSL2
|
ADAMTS-like 2
|
|
chr2_+_173600519
|
1.032
|
NM_007023
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr19_+_40146533
|
1.032
|
NM_001190441
|
LGALS16
|
lectin, galactoside-binding, soluble, 16
|
|
chr11_-_65314049
|
1.031
|
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr12_-_6484557
|
1.027
|
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr18_-_31803168
|
1.027
|
|
NOL4
|
nucleolar protein 4
|
|
chr8_+_144816287
|
1.025
|
|
LOC100128338
|
uncharacterized LOC100128338
|
|
chr17_+_1958354
|
1.022
|
NM_006497
|
HIC1
|
hypermethylated in cancer 1
|
|
chr4_-_25032306
|
1.018
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr21_-_38639593
|
1.018
|
|
DSCR3
|
Down syndrome critical region gene 3
|
|
chr1_+_183155393
|
1.014
|
|
LAMC2
|
laminin, gamma 2
|
|
chr1_-_22109669
|
1.010
|
NM_001032730
NM_032236
|
USP48
|
ubiquitin specific peptidase 48
|
|
chr16_+_127017
|
1.009
|
NM_001015054
|
MPG
|
N-methylpurine-DNA glycosylase
|
|
chr19_+_54385429
|
1.003
|
NM_002739
|
PRKCG
|
protein kinase C, gamma
|
|
chr20_-_23030141
|
1.003
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr14_+_105190533
|
1.000
|
NM_152328
|
ADSSL1
|
adenylosuccinate synthase like 1
|
|
chr22_-_19512001
|
1.000
|
|
CLDN5
|
claudin 5
|
|
chr3_-_178789535
|
1.000
|
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr11_-_59612925
|
0.995
|
NM_005142
|
GIF
|
gastric intrinsic factor (vitamin B synthesis)
|
|
chr7_-_155604815
|
0.994
|
NM_000193
|
SHH
|
sonic hedgehog
|
|
chr14_-_106877689
|
0.993
|
|
IGHV4-31
IGH@
IGHA1
IGHA2
IGHG1
IGHG4
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy locus
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 4 (G4m marker)
immunoglobulin heavy constant mu
|
|
chr7_+_1580009
|
0.992
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr1_-_32707198
|
0.992
|
|
MTMR9LP
|
myotubularin related protein 9-like, pseudogene
|
|
chr5_-_134787987
|
0.990
|
NM_001099221
|
TIFAB
|
TRAF-interacting protein with forkhead-associated domain, family member B
|
|
chr21_+_30517897
|
0.985
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr17_-_76921205
|
0.984
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr20_+_44044706
|
0.984
|
NM_001184728
NM_001184729
NM_001184730
NM_015937
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis, class T
|
|
chr15_-_38856830
|
0.983
|
NM_001128602
NM_005739
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated)
|