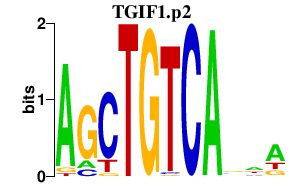
|
chr13_-_60325073
|
2.491
|
|
|
|
|
chr1_-_24469571
|
1.994
|
NM_021258
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr4_+_69313166
|
1.896
|
NM_014058
|
TMPRSS11E
|
transmembrane protease, serine 11E
|
|
chr1_+_17531619
|
1.805
|
NM_013358
|
PADI1
|
peptidyl arginine deiminase, type I
|
|
chr1_-_152386727
|
1.753
|
NM_016190
|
CRNN
|
cornulin
|
|
chr17_-_39092714
|
1.495
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr12_+_53442732
|
1.477
|
NM_015319
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr9_+_125133322
|
1.469
|
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr2_-_113594325
|
1.339
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr22_-_37640187
|
1.303
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr4_-_175443601
|
1.300
|
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD)
|
|
chr6_-_41130820
|
1.273
|
|
TREM2
|
triggering receptor expressed on myeloid cells 2
|
|
chr4_-_175443791
|
1.263
|
NM_000860
NM_001145816
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD)
|
|
chr1_+_32043745
|
1.174
|
NM_001204415
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr6_-_41130864
|
1.122
|
|
TREM2
|
triggering receptor expressed on myeloid cells 2
|
|
chr9_+_125133228
|
1.103
|
NM_000962
NM_080591
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr9_+_34652125
|
1.101
|
NM_001142784
|
IL11RA
|
interleukin 11 receptor, alpha
|
|
chr3_-_189840225
|
1.100
|
NM_001134418
|
LEPREL1
|
leprecan-like 1
|
|
chr16_+_69139466
|
1.088
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr10_+_102123420
|
1.076
|
|
|
|
|
chr5_+_147691988
|
1.072
|
NM_032566
|
SPINK7
|
serine peptidase inhibitor, Kazal type 7 (putative)
|
|
chr17_+_57232859
|
1.059
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr13_+_78109808
|
1.053
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr14_-_24732344
|
1.045
|
NM_000359
|
TGM1
|
transglutaminase 1 (K polypeptide epidermal type I, protein-glutamine-gamma-glutamyltransferase)
|
|
chr22_-_37640287
|
1.038
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr1_-_205391180
|
1.037
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr1_+_153388999
|
1.034
|
NM_176823
|
S100A7A
|
S100 calcium binding protein A7A
|
|
chr4_+_88896838
|
1.019
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr3_-_127541975
|
1.012
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr17_+_76210329
|
1.000
|
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr4_+_88896801
|
0.995
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr2_+_27719702
|
0.988
|
NM_001486
|
GCKR
|
glucokinase (hexokinase 4) regulator
|
|
chr4_+_88896899
|
0.974
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr4_+_88896868
|
0.957
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr6_-_41130921
|
0.949
|
NM_018965
|
TREM2
|
triggering receptor expressed on myeloid cells 2
|
|
chrX_-_153775427
|
0.945
|
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr9_+_116207010
|
0.930
|
NM_144488
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_-_72287097
|
0.893
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr10_+_94352960
|
0.831
|
|
KIF11
|
kinesin family member 11
|
|
chr2_+_239756672
|
0.828
|
NM_057179
|
TWIST2
|
twist homolog 2 (Drosophila)
|
|
chr22_+_44395090
|
0.811
|
NM_001003828
|
PARVB
|
parvin, beta
|
|
chr17_+_4402128
|
0.790
|
NM_001124758
|
SPNS2
|
spinster homolog 2 (Drosophila)
|
|
chr12_+_107078476
|
0.780
|
NM_032491
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chr12_-_117747214
|
0.776
|
NM_001204214
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr3_-_127541160
|
0.773
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr18_+_61420276
|
0.773
|
NM_001040147
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr2_-_216003068
|
0.761
|
NM_173076
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chrX_-_38079980
|
0.761
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr11_+_129245834
|
0.757
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr1_+_156252725
|
0.753
|
|
TMEM79
|
transmembrane protein 79
|
|
chr2_+_233925104
|
0.746
|
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr1_-_153433104
|
0.727
|
NM_002963
|
S100A7
|
S100 calcium binding protein A7
|
|
chr19_-_36643770
|
0.727
|
NM_001864
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle)
|
|
chr17_-_39728304
|
0.724
|
NM_000226
|
KRT9
|
keratin 9
|
|
chr19_-_14228560
|
0.723
|
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr5_-_127873734
|
0.715
|
NM_001999
|
FBN2
|
fibrillin 2
|
|
chr6_+_80714389
|
0.710
|
|
TTK
|
TTK protein kinase
|
|
chr1_+_24645914
|
0.694
|
NM_001195010
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr6_+_80714321
|
0.690
|
NM_001166691
NM_003318
|
TTK
|
TTK protein kinase
|
|
chr12_+_15475190
|
0.686
|
NM_002848
NM_030667
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chrX_+_70443055
|
0.682
|
NM_000166
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr17_-_39768939
|
0.680
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr22_-_30234253
|
0.672
|
NM_001242906
NM_032204
|
ASCC2
|
activating signal cointegrator 1 complex subunit 2
|
|
chr17_-_39661803
|
0.656
|
NM_002274
NM_153490
|
KRT13
|
keratin 13
|
|
chr2_+_238536183
|
0.656
|
NM_001137550
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr11_-_123756339
|
0.638
|
NM_001013743
|
TMEM225
|
transmembrane protein 225
|
|
chr1_-_150978879
|
0.637
|
NM_001163259
NM_001163260
NM_001040217
|
FAM63A
|
family with sequence similarity 63, member A
|
|
chr17_-_39525873
|
0.633
|
NM_002279
|
KRT33B
|
keratin 33B
|
|
chr1_+_156254069
|
0.633
|
NM_032323
|
TMEM79
|
transmembrane protein 79
|
|
chr2_+_33359607
|
0.631
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr22_-_50523853
|
0.629
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr2_+_233925035
|
0.625
|
NM_001017915
NM_005541
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr6_-_41863086
|
0.621
|
NM_018561
|
USP49
|
ubiquitin specific peptidase 49
|
|
chr19_-_44285407
|
0.602
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr12_+_53497287
|
0.601
|
|
SOAT2
|
sterol O-acyltransferase 2
|
|
chr5_-_127873477
|
0.596
|
|
FBN2
|
fibrillin 2
|
|
chr10_-_90750955
|
0.594
|
NM_001141945
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr19_-_41934634
|
0.585
|
NM_198540
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8
|
|
chr14_-_107078578
|
0.585
|
|
|
|
|
chr7_+_139528951
|
0.582
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr9_-_36276930
|
0.581
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr4_+_5526882
|
0.580
|
NM_005750
|
C4orf6
|
chromosome 4 open reading frame 6
|
|
chr18_-_67872897
|
0.579
|
NM_173630
|
RTTN
|
rotatin
|
|
chr11_-_74442185
|
0.578
|
NM_015424
|
CHRDL2
|
chordin-like 2
|
|
chr8_-_144654921
|
0.577
|
NM_001100878
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr9_+_35673852
|
0.574
|
NM_001216
|
CA9
|
carbonic anhydrase IX
|
|
chr4_-_47983430
|
0.571
|
NM_001142564
|
CNGA1
|
cyclic nucleotide gated channel alpha 1
|
|
chr7_+_77469446
|
0.569
|
NM_001127357
NM_001127359
NM_020432
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr9_+_34990715
|
0.568
|
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr19_+_45409003
|
0.564
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr13_+_28494136
|
0.563
|
NM_000209
|
PDX1
|
pancreatic and duodenal homeobox 1
|
|
chr11_+_842821
|
0.562
|
NM_001025238
NM_001025239
NM_003271
NM_001025235
NM_001025236
|
TSPAN4
|
tetraspanin 4
|
|
chrX_-_153775774
|
0.552
|
NM_001042351
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr10_+_124220990
|
0.541
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr19_-_49865638
|
0.541
|
NM_003598
|
TEAD2
|
TEA domain family member 2
|
|
chr5_-_149682447
|
0.540
|
NM_001012301
|
ARSI
|
arylsulfatase family, member I
|
|
chr11_-_2018568
|
0.540
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr4_-_143767603
|
0.537
|
NM_001101669
NM_003866
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa
|
|
chr11_-_568419
|
0.532
|
|
MIR210HG
|
MIR210 host gene (non-protein coding)
|
|
chr1_-_169599349
|
0.532
|
NM_003005
|
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62)
|
|
chr4_-_114682340
|
0.529
|
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta
|
|
chr16_+_71392615
|
0.527
|
NM_001740
NM_007088
|
CALB2
|
calbindin 2
|
|
chr16_-_3030473
|
0.523
|
NM_004203
NM_182687
|
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1
|
|
chr16_+_71392637
|
0.521
|
|
CALB2
|
calbindin 2
|
|
chr11_-_119187811
|
0.521
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr22_+_31487315
|
0.520
|
|
SMTN
|
smoothelin
|
|
chr1_-_208417635
|
0.517
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr19_-_44285012
|
0.513
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr4_-_156787377
|
0.512
|
NM_017419
|
ACCN5
|
amiloride-sensitive cation channel 5, intestinal
|
|
chr6_+_24775154
|
0.510
|
NM_001251989
NM_001251990
NM_015895
|
GMNN
|
geminin, DNA replication inhibitor
|
|
chr1_+_6615337
|
0.504
|
NM_138697
NM_177540
|
TAS1R1
|
taste receptor, type 1, member 1
|
|
chr12_+_53497273
|
0.501
|
NM_003578
|
SOAT2
|
sterol O-acyltransferase 2
|
|
chr11_-_64825992
|
0.499
|
NM_005468
|
NAALADL1
|
N-acetylated alpha-linked acidic dipeptidase-like 1
|
|
chr12_+_56325788
|
0.496
|
NM_201444
NM_201445
NM_201554
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr12_+_119616594
|
0.493
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr8_-_125580745
|
0.489
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr3_-_9595412
|
0.489
|
NM_198560
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4
|
|
chr20_+_20037289
|
0.484
|
NM_001167816
|
C20orf26
|
chromosome 20 open reading frame 26
|
|
chr10_-_88853826
|
0.478
|
|
GLUD1
|
glutamate dehydrogenase 1
|
|
chr4_-_82126200
|
0.476
|
NM_006259
|
PRKG2
|
protein kinase, cGMP-dependent, type II
|
|
chrX_+_101380659
|
0.475
|
NM_080390
|
TCEAL2
|
transcription elongation factor A (SII)-like 2
|
|
chr16_-_67217403
|
0.469
|
|
KIAA0895L
|
KIAA0895-like
|
|
chr1_+_24645811
|
0.467
|
NM_198173
NM_198174
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr14_+_24600674
|
0.466
|
NM_203402
|
FITM1
|
fat storage-inducing transmembrane protein 1
|
|
chr1_+_207002221
|
0.465
|
NM_013371
|
IL19
|
interleukin 19
|
|
chr1_+_74701070
|
0.463
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr19_-_39826676
|
0.462
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr4_+_55095263
|
0.461
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr22_-_50523737
|
0.459
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr8_+_28174648
|
0.459
|
NM_006228
|
PNOC
|
prepronociceptin
|
|
chr22_-_50523780
|
0.458
|
NM_139202
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr22_+_46692637
|
0.457
|
NM_016426
|
GTSE1
|
G-2 and S-phase expressed 1
|
|
chr6_+_24775640
|
0.457
|
NM_001251991
|
GMNN
|
geminin, DNA replication inhibitor
|
|
chr7_+_141490016
|
0.456
|
NM_018980
|
TAS2R5
|
taste receptor, type 2, member 5
|
|
chr16_-_67217825
|
0.453
|
NM_001040715
|
KIAA0895L
|
KIAA0895-like
|
|
chr17_+_16945766
|
0.453
|
NM_015134
NM_201274
|
MPRIP
|
myosin phosphatase Rho interacting protein
|
|
chr2_+_220436953
|
0.452
|
NM_002191
|
INHA
|
inhibin, alpha
|
|
chr4_-_114682594
|
0.451
|
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta
|
|
chr5_-_95748688
|
0.451
|
NM_001177876
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr1_-_11907741
|
0.446
|
NM_006172
|
NPPA
|
natriuretic peptide A
|
|
chr5_-_112824526
|
0.445
|
NM_001085377
|
MCC
|
mutated in colorectal cancers
|
|
chr7_-_27192201
|
0.444
|
|
|
|
|
chr2_+_33661392
|
0.443
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr9_-_123639462
|
0.441
|
|
PHF19
|
PHD finger protein 19
|
|
chr2_+_233243243
|
0.440
|
NM_001632
|
ALPP
|
alkaline phosphatase, placental
|
|
chr2_+_211421322
|
0.439
|
NM_001875
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr11_+_2924504
|
0.433
|
|
SLC22A18
|
solute carrier family 22, member 18
|
|
chr22_+_25615611
|
0.432
|
NM_000496
|
CRYBB2
|
crystallin, beta B2
|
|
chr5_+_115298136
|
0.432
|
NM_173800
|
AQPEP
|
laeverin
|
|
chr17_-_57232799
|
0.425
|
NM_001100595
NM_182620
|
SKA2
|
spindle and kinetochore associated complex subunit 2
|
|
chr4_+_186990308
|
0.421
|
NM_003265
|
TLR3
|
toll-like receptor 3
|
|
chr16_+_69141442
|
0.418
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr9_+_706744
|
0.412
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr17_+_57233100
|
0.411
|
|
PRR11
|
proline rich 11
|
|
chr17_-_39023461
|
0.409
|
NM_000223
|
KRT12
|
keratin 12
|
|
chr3_-_48632592
|
0.408
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr8_+_38243958
|
0.407
|
NM_001199659
NM_144652
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2
|
|
chr9_+_128510477
|
0.406
|
NM_001134778
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr17_-_34122547
|
0.401
|
NM_001032278
NM_024302
|
MMP28
|
matrix metallopeptidase 28
|
|
chrX_+_41306686
|
0.391
|
NM_022567
|
NYX
|
nyctalopin
|
|
chr9_+_2622134
|
0.387
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr9_-_132805456
|
0.384
|
NM_015033
|
FNBP1
|
formin binding protein 1
|
|
chr4_+_147560042
|
0.384
|
NM_004575
|
POU4F2
|
POU class 4 homeobox 2
|
|
chr20_+_33759773
|
0.384
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr9_+_125132823
|
0.377
|
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr10_-_14372807
|
0.377
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr10_+_115310589
|
0.375
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr22_-_50524357
|
0.375
|
NM_015166
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr6_-_37225360
|
0.371
|
NM_145316
|
TMEM217
|
transmembrane protein 217
|
|
chr9_+_2622099
|
0.369
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr15_+_63336274
|
0.368
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr9_+_2622052
|
0.367
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr8_-_124408688
|
0.366
|
NM_014109
|
ATAD2
|
ATPase family, AAA domain containing 2
|
|
chr4_-_176923572
|
0.366
|
NM_005277
NM_201592
|
GPM6A
|
glycoprotein M6A
|
|
chr4_-_185747051
|
0.366
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr2_+_234668914
|
0.360
|
NM_000463
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1
|
|
chr17_-_7493389
|
0.355
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr18_+_61144143
|
0.353
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr14_-_100842637
|
0.351
|
|
WARS
|
tryptophanyl-tRNA synthetase
|
|
chr15_-_42840992
|
0.349
|
NM_153260
|
LRRC57
|
leucine rich repeat containing 57
|
|
chr6_+_151186884
|
0.347
|
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr6_-_131384387
|
0.346
|
NM_001135554
NM_001135555
NM_001199388
NM_001431
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr3_+_113251217
|
0.344
|
NM_017699
|
SIDT1
|
SID1 transmembrane family, member 1
|
|
chr17_-_3499124
|
0.343
|
NM_080705
|
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1
|
|
chr12_-_53074172
|
0.341
|
NM_006121
|
KRT1
|
keratin 1
|
|
chr7_+_76139744
|
0.340
|
NM_030570
NM_182684
|
UPK3B
|
uroplakin 3B
|
|
chr6_+_31674639
|
0.340
|
NM_001003693
|
LY6G6F
|
lymphocyte antigen 6 complex, locus G6F
|
|
chr8_-_49833823
|
0.337
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr17_-_57232524
|
0.334
|
|
SKA2
|
spindle and kinetochore associated complex subunit 2
|
|
chr19_-_11565612
|
0.331
|
|
|
|
|
chr10_+_91498724
|
0.331
|
|
KIF20B
|
kinesin family member 20B
|
|
chr19_-_48547180
|
0.331
|
NM_019855
|
CABP5
|
calcium binding protein 5
|
|
chr7_-_99766191
|
0.329
|
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr12_+_13043955
|
0.326
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr20_+_43344005
|
0.326
|
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr7_-_99766238
|
0.325
|
NM_024637
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr13_-_34250899
|
0.324
|
NM_001243476
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr2_-_119605758
|
0.322
|
NM_001426
|
EN1
|
engrailed homeobox 1
|
|
chr1_-_42384370
|
0.320
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr19_-_39826625
|
0.318
|
|
GMFG
|
glia maturation factor, gamma
|