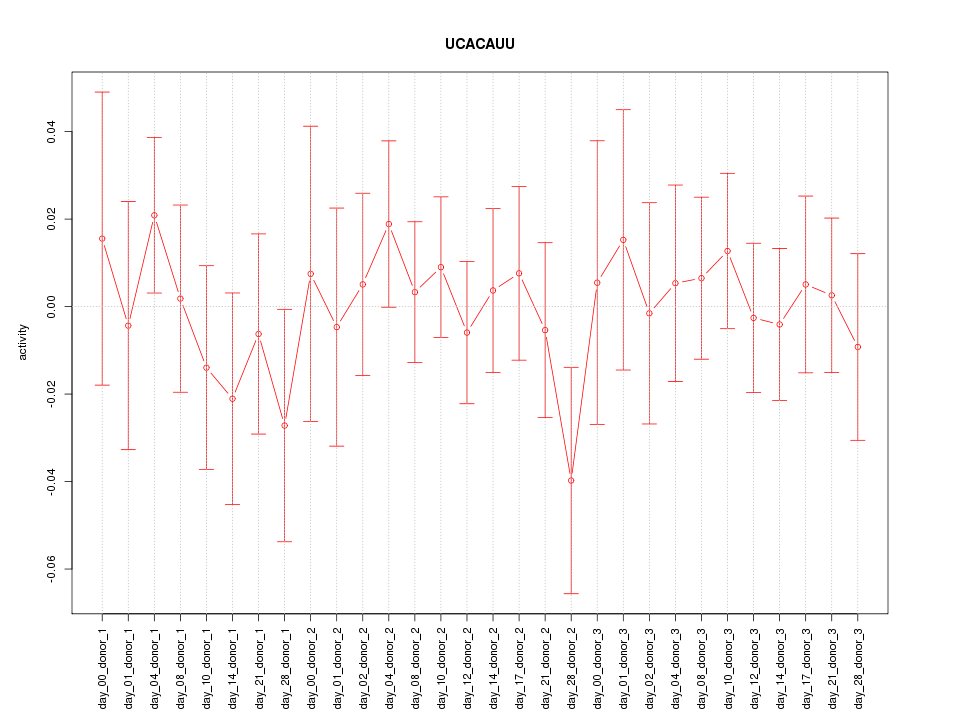
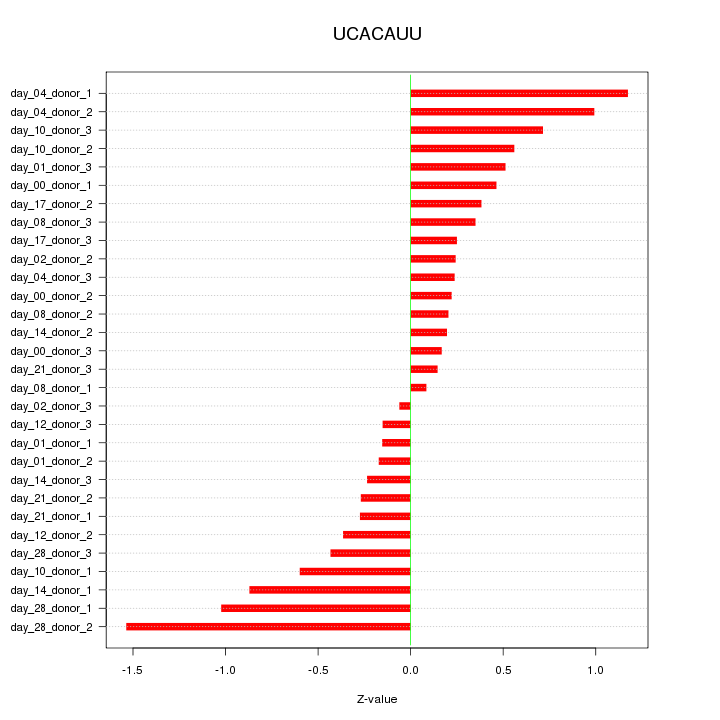
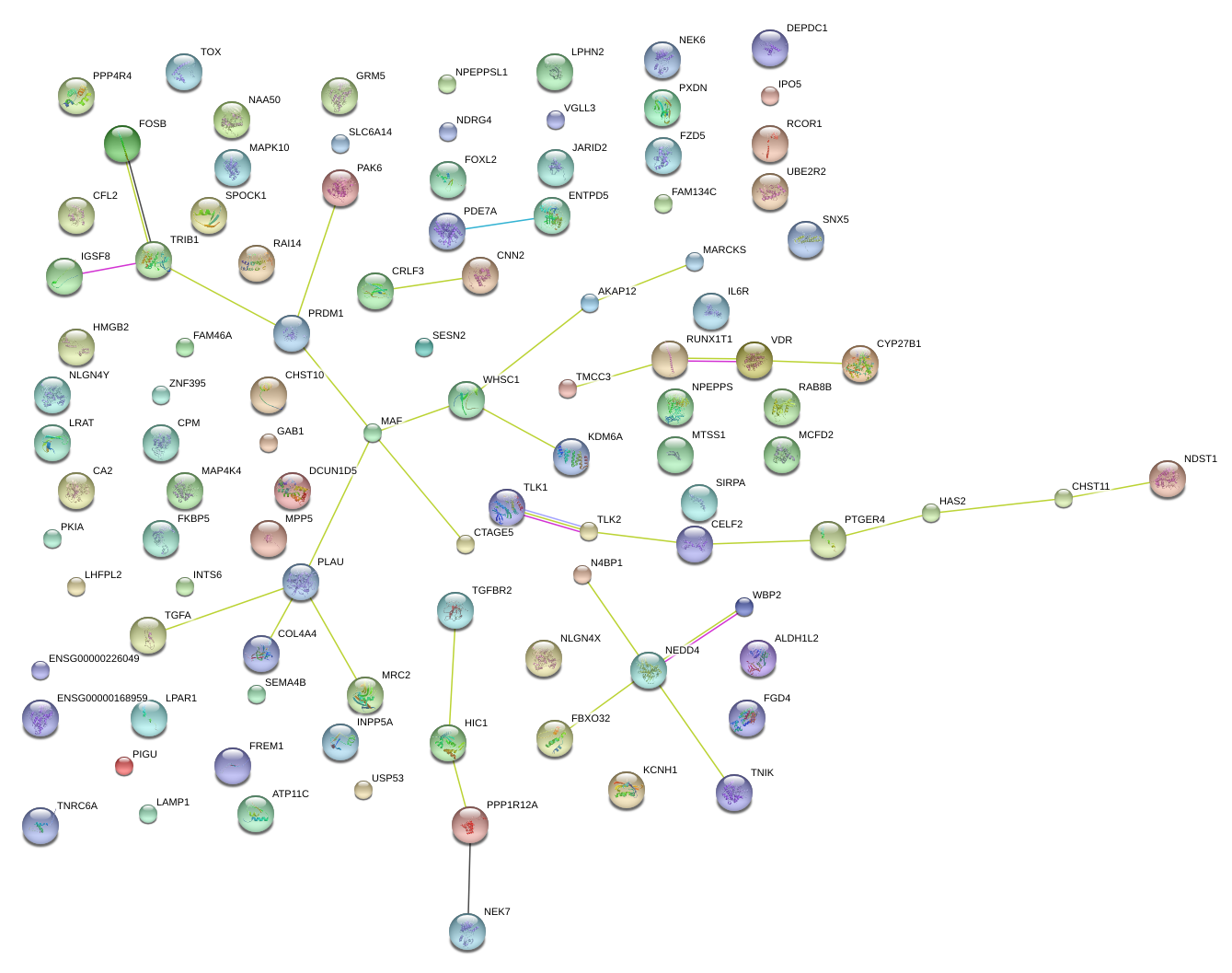
|
chr5_-_136834887
|
1.872
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr2_-_1748285
|
1.370
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr6_+_151646634
|
1.038
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr15_+_90728118
|
1.025
|
NM_020210
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr6_-_35696359
|
1.025
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr6_+_151561094
|
0.911
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr6_-_35656677
|
0.807
|
NM_001145776
NM_001145777
NM_004117
|
FKBP5
|
FK506 binding protein 5
|
|
chr15_+_90744506
|
0.733
|
NM_198925
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr20_+_1875382
|
0.627
|
NM_080792
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr20_+_1875825
|
0.621
|
NM_001040023
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr20_+_1874779
|
0.597
|
NM_001040022
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr2_+_102314162
|
0.582
|
NM_001242560
NM_004834
NM_145686
NM_001242559
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr5_+_34684611
|
0.560
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chrX_+_115567746
|
0.557
|
NM_007231
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr8_+_86376130
|
0.556
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr17_+_60704747
|
0.537
|
NM_006039
|
MRC2
|
mannose receptor, C type 2
|
|
chr15_+_63481713
|
0.500
|
NM_016530
|
RAB8B
|
RAB8B, member RAS oncogene family
|
|
chr8_-_28243941
|
0.477
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr10_+_11206928
|
0.473
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr9_-_113800243
|
0.471
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr5_+_34687663
|
0.471
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr5_-_77944341
|
0.448
|
NM_005779
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2
|
|
chr4_+_1873035
|
0.446
|
NM_001042424
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr19_+_1026271
|
0.427
|
NM_004368
NM_201277
|
CNN2
|
calponin 2
|
|
chr13_+_98605928
|
0.421
|
NM_002271
|
IPO5
|
importin 5
|
|
chr12_+_32654976
|
0.415
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr9_+_127054246
|
0.391
|
NM_001166170
NM_001166171
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr4_+_1894508
|
0.391
|
NM_007331
NM_133330
NM_133331
NM_133335
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr9_+_127054567
|
0.389
|
NM_001166169
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr10_+_11047253
|
0.388
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr6_+_15246299
|
0.381
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr11_-_88796815
|
0.373
|
NM_000842
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr5_+_34656595
|
0.361
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr5_+_149887673
|
0.345
|
NM_001543
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr5_+_34656367
|
0.336
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr2_-_208634051
|
0.335
|
NM_003468
|
FZD5
|
frizzled family receptor 5
|
|
chr11_-_88781039
|
0.335
|
NM_001143831
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr10_+_11059854
|
0.334
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr3_-_87040246
|
0.328
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr8_+_126442557
|
0.320
|
NM_025195
|
TRIB1
|
tribbles homolog 1 (Drosophila)
|
|
chr1_+_154377668
|
0.314
|
NM_000565
NM_001206866
NM_181359
|
IL6R
|
interleukin 6 receptor
|
|
chr14_+_103058988
|
0.313
|
NM_015156
|
RCOR1
|
REST corepressor 1
|
|
chr5_+_40680021
|
0.303
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr9_+_127020462
|
0.301
|
NM_001166168
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr4_+_144257982
|
0.298
|
NM_002039
NM_207123
|
GAB1
|
GRB2-associated binding protein 1
|
|
chr3_-_113465101
|
0.291
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr15_-_56209305
|
0.290
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr6_+_114178516
|
0.290
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chrX_-_138914384
|
0.288
|
NM_001010986
NM_173694
|
ATP11C
|
ATPase, class VI, type 11C
|
|
chr9_+_127019782
|
0.284
|
NM_001166167
NM_001145001
NM_014397
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr1_+_28585959
|
0.284
|
NM_031459
|
SESN2
|
sestrin 2
|
|
chr2_-_47143250
|
0.280
|
NM_001171507
NM_001171510
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr8_-_60031545
|
0.280
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr8_-_66701175
|
0.277
|
NM_002603
|
PDE7A
|
phosphodiesterase 7A
|
|
chr12_-_80307690
|
0.274
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr1_-_211307314
|
0.272
|
NM_002238
NM_172362
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1
|
|
chr17_+_1958354
|
0.272
|
NM_006497
|
HIC1
|
hypermethylated in cancer 1
|
|
chr19_+_45971249
|
0.270
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr2_-_101034049
|
0.264
|
NM_004854
|
CHST10
|
carbohydrate sulfotransferase 10
|
|
chr12_-_48298765
|
0.263
|
NM_000376
NM_001017535
NM_001017536
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor
|
|
chr16_-_79634610
|
0.263
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chrX_-_6145887
|
0.257
|
NM_020742
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr12_-_95044205
|
0.256
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr15_-_56285732
|
0.253
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr12_-_69357019
|
0.252
|
NM_001874
|
CPM
|
carboxypeptidase M
|
|
chr2_-_47142950
|
0.250
|
NM_001171506
NM_001171509
NM_139279
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr2_-_172087823
|
0.247
|
NM_001136554
|
TLK1
|
tousled-like kinase 1
|
|
chr2_-_171923482
|
0.244
|
NM_001136555
|
TLK1
|
tousled-like kinase 1
|
|
chr17_-_73851377
|
0.240
|
NM_012478
|
WBP2
|
WW domain binding protein 2
|
|
chr17_+_1959512
|
0.240
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr8_-_93075190
|
0.238
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_47168902
|
0.235
|
NM_001171508
NM_001171511
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chrX_-_6146655
|
0.234
|
NM_181332
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr12_-_69326946
|
0.234
|
NM_001005502
NM_198320
|
CPM
|
carboxypeptidase M
|
|
chr1_-_68962726
|
0.233
|
NM_001114120
NM_017779
|
DEPDC1
|
DEP domain containing 1
|
|
chr12_-_80329234
|
0.229
|
NM_001143885
NM_001244990
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr14_+_94640648
|
0.224
|
NM_020958
NM_058237
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4
|
|
chr8_+_79428335
|
0.224
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr1_+_82266081
|
0.222
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr3_-_138665981
|
0.216
|
NM_023067
|
FOXL2
|
forkhead box L2
|
|
chr8_-_124544376
|
0.214
|
NM_148177
|
FBXO32
|
F-box protein 32
|
|
chr16_-_48644087
|
0.206
|
NM_153029
|
N4BP1
|
NEDD4 binding protein 1
|
|
chr12_+_104850641
|
0.205
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr4_+_120133764
|
0.204
|
NM_019050
|
USP53
|
ubiquitin specific peptidase 53
|
|
chr12_-_80328926
|
0.203
|
NM_002480
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr9_-_14910992
|
0.202
|
NM_144966
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr17_+_45608443
|
0.202
|
NM_006310
|
NPEPPS
|
aminopeptidase puromycin sensitive
|
|
chr2_-_70781146
|
0.199
|
NM_001099691
NM_003236
|
TGFA
|
transforming growth factor, alpha
|
|
chr10_+_134351272
|
0.198
|
NM_005539
|
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa
|
|
chr20_-_17949384
|
0.197
|
NM_152227
|
SNX5
|
sorting nexin 5
|
|
chr6_-_82462374
|
0.197
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr4_+_166248816
|
0.196
|
NM_001017369
NM_006745
|
MSMO1
|
methylsterol monooxygenase 1
|
|
chr2_-_172017358
|
0.196
|
NM_012290
|
TLK1
|
tousled-like kinase 1
|
|
chr6_+_106534188
|
0.194
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr14_-_35182944
|
0.193
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr13_+_113951461
|
0.191
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr9_+_33817168
|
0.188
|
NM_017811
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2
|
|
chr15_+_40509628
|
0.188
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr11_-_102962903
|
0.188
|
NM_032299
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae)
|
|
chr14_+_39734475
|
0.187
|
NM_203354
|
CTAGE5
|
CTAGE family, member 5
|
|
chr9_-_14779530
|
0.182
|
NM_001177704
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr8_-_93029864
|
0.182
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_160068617
|
0.182
|
NM_001206665
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chr14_-_74485813
|
0.182
|
NM_001249
|
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5
|
|
chr14_-_35183722
|
0.179
|
NM_001243645
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr12_-_105478218
|
0.179
|
NM_001034173
|
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2
|
|
chr8_-_124553448
|
0.177
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr10_+_75670861
|
0.176
|
NM_001145031
NM_002658
|
PLAU
|
plasminogen activator, urokinase
|
|
chr8_-_93111915
|
0.176
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_160068359
|
0.172
|
NM_052868
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chr13_-_52027274
|
0.169
|
NM_001039937
NM_001039938
NM_012141
|
INTS6
|
integrator complex subunit 6
|
|
chr17_-_40761404
|
0.168
|
NM_178126
|
FAM134C
|
family with sequence similarity 134, member C
|
|
chr16_+_58498725
|
0.167
|
NM_022910
|
NDRG4
|
NDRG family member 4
|
|
chr14_+_67707825
|
0.164
|
NM_022474
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5)
|
|
chr4_+_155665150
|
0.163
|
NM_004744
|
LRAT
|
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase)
|
|
chr8_-_122653493
|
0.161
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr16_+_58497548
|
0.161
|
NM_020465
|
NDRG4
|
NDRG family member 4
|
|
chr2_-_228029274
|
0.160
|
NM_000092
|
COL4A4
|
collagen, type IV, alpha 4
|
|
chr14_+_39736277
|
0.160
|
NM_001247989
NM_001247990
NM_005930
NM_203355
|
CTAGE5
|
CTAGE family, member 5
|
|
chr4_-_174254730
|
0.158
|
NM_001130689
|
HMGB2
|
high mobility group box 2
|
|
chr16_+_24740945
|
0.158
|
NM_014494
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr3_+_30647901
|
0.157
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr16_+_58497993
|
0.156
|
NM_001130487
|
NDRG4
|
NDRG family member 4
|
|
chr8_-_125740650
|
0.154
|
NM_014751
|
MTSS1
|
metastasis suppressor 1
|
|
chr17_-_29151724
|
0.151
|
NM_015986
|
CRLF3
|
cytokine receptor-like factor 3
|
|
chrX_+_44732402
|
0.148
|
NM_021140
|
KDM6A
|
lysine (K)-specific demethylase 6A
|
|
chrX_-_30907408
|
0.148
|
NM_152787
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3
|
|
chr12_+_123868608
|
0.147
|
NM_020382
|
SETD8
|
SET domain containing (lysine methyltransferase) 8
|
|
chr16_+_27438578
|
0.146
|
NM_021798
|
IL21R
|
interleukin 21 receptor
|
|
chr15_+_83477972
|
0.146
|
NM_001080435
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules
|
|
chr6_+_138188352
|
0.145
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr1_+_46972667
|
0.144
|
NM_147192
NM_172225
|
DMBX1
|
diencephalon/mesencephalon homeobox 1
|
|
chr10_+_60094712
|
0.143
|
NM_001204880
NM_003338
|
UBE2D1
|
ubiquitin-conjugating enzyme E2D 1
|
|
chr18_-_48724031
|
0.143
|
NM_016626
|
MEX3C
|
mex-3 homolog C (C. elegans)
|
|
chr6_+_106546736
|
0.141
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr1_-_154155698
|
0.141
|
NM_001043351
NM_001043352
NM_001043353
NM_153649
|
TPM3
|
tropomyosin 3
|
|
chr2_-_37193609
|
0.140
|
NM_003162
|
STRN
|
striatin, calmodulin binding protein
|
|
chr11_+_133938819
|
0.139
|
NM_001205329
NM_032801
|
JAM3
|
junctional adhesion molecule 3
|
|
chr3_+_147127151
|
0.138
|
NM_003412
|
ZIC1
|
Zic family member 1
|
|
chr14_+_37126772
|
0.138
|
NM_006194
|
PAX9
|
paired box 9
|
|
chr8_-_55014367
|
0.137
|
NM_006330
|
LYPLA1
|
lysophospholipase I
|
|
chr1_+_53192021
|
0.135
|
NM_024646
|
ZYG11B
|
zyg-11 homolog B (C. elegans)
|
|
chr1_+_23037329
|
0.134
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr20_+_43595119
|
0.133
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr13_-_30169795
|
0.133
|
NM_003045
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1
|
|
chr15_-_30114536
|
0.133
|
NM_003257
NM_175610
|
TJP1
|
tight junction protein 1 (zona occludens 1)
|
|
chr1_-_231560789
|
0.131
|
NM_022051
|
EGLN1
|
egl nine homolog 1 (C. elegans)
|
|
chr6_+_121756744
|
0.130
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr9_-_19102898
|
0.130
|
NM_017645
|
HAUS6
|
HAUS augmin-like complex, subunit 6
|
|
chr12_+_104324186
|
0.129
|
NM_003299
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chrX_-_129244471
|
0.129
|
NM_001421
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr18_-_31628557
|
0.128
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chrX_-_41782230
|
0.127
|
NM_001126054
NM_001126055
NM_003688
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family)
|
|
chrX_-_129244652
|
0.126
|
NM_001127197
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr9_+_4490193
|
0.124
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr2_+_191745546
|
0.121
|
NM_014905
|
GLS
|
glutaminase
|
|
chr17_+_61086897
|
0.121
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr18_+_9913954
|
0.121
|
NM_003574
NM_194434
|
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa
|
|
chr9_+_129567260
|
0.120
|
NM_001135776
NM_014007
|
ZBTB43
|
zinc finger and BTB domain containing 43
|
|
chr11_+_69455837
|
0.120
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr7_-_14030864
|
0.119
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chr3_-_98620452
|
0.117
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr7_-_14031049
|
0.116
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr17_-_16472463
|
0.116
|
NM_020653
|
ZNF287
|
zinc finger protein 287
|
|
chr11_+_86748870
|
0.114
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr9_-_95527002
|
0.112
|
NM_001003800
NM_015250
|
BICD2
|
bicaudal D homolog 2 (Drosophila)
|
|
chr6_+_105404922
|
0.112
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr5_-_148442606
|
0.112
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr4_+_15004297
|
0.112
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr18_-_31803514
|
0.112
|
NM_001198546
NM_001198548
NM_003787
|
NOL4
|
nucleolar protein 4
|
|
chr16_+_58534045
|
0.109
|
NM_001242833
NM_001242834
NM_001242835
NM_001242836
|
NDRG4
|
NDRG family member 4
|
|
chr9_-_95432542
|
0.109
|
NM_022755
|
IPPK
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase
|
|
chr13_-_80915041
|
0.108
|
NM_005842
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr20_-_17949067
|
0.108
|
NM_014426
|
SNX5
|
sorting nexin 5
|
|
chr18_-_31802433
|
0.107
|
NM_001198547
|
NOL4
|
nucleolar protein 4
|
|
chr17_+_78518611
|
0.107
|
NM_001163034
NM_020761
|
RPTOR
|
regulatory associated protein of MTOR, complex 1
|
|
chr14_+_55518313
|
0.106
|
NM_144578
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like
|
|
chr1_-_240775448
|
0.104
|
NM_022469
|
GREM2
|
gremlin 2
|
|
chr12_-_54653369
|
0.104
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr16_-_15982446
|
0.104
|
NM_144600
|
FOPNL
|
FGFR1OP N-terminal like
|
|
chr6_+_96463844
|
0.103
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr7_+_4721722
|
0.103
|
NM_001037165
|
FOXK1
|
forkhead box K1
|
|
chr9_+_110045516
|
0.102
|
NM_002874
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr4_-_174255454
|
0.101
|
NM_001130688
NM_002129
|
HMGB2
|
high mobility group box 2
|
|
chr5_-_78282356
|
0.101
|
NM_000046
|
ARSB
|
arylsulfatase B
|
|
chrX_-_63425484
|
0.100
|
NM_152424
|
FAM123B
|
family with sequence similarity 123B
|
|
chr7_+_38217807
|
0.100
|
NM_032016
|
STARD3NL
|
STARD3 N-terminal like
|
|
chr1_-_27816676
|
0.100
|
NM_001201404
NM_006990
|
WASF2
|
WAS protein family, member 2
|
|
chr14_+_102786083
|
0.099
|
NM_018335
|
ZNF839
|
zinc finger protein 839
|
|
chr3_-_71778268
|
0.099
|
NM_001134650
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr1_+_90098643
|
0.098
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr5_+_72112417
|
0.098
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr8_+_27179918
|
0.098
|
NM_004103
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr12_-_46663207
|
0.097
|
NM_001077484
NM_030674
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr15_+_67358194
|
0.097
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr8_-_66753968
|
0.096
|
NM_001242318
|
PDE7A
|
phosphodiesterase 7A
|
|
chr2_+_28974608
|
0.096
|
NM_206876
NM_002709
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme
|
|
chr8_-_131455833
|
0.095
|
NM_001247996
NM_018482
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1
|
|
chr1_-_214724974
|
0.095
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr8_+_77593506
|
0.094
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|