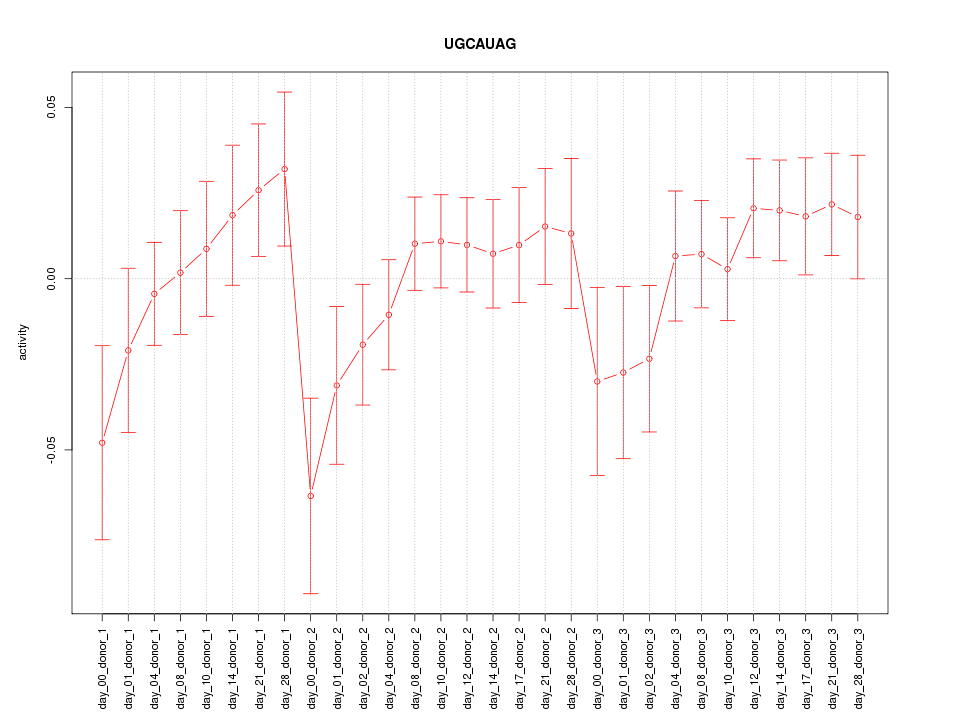
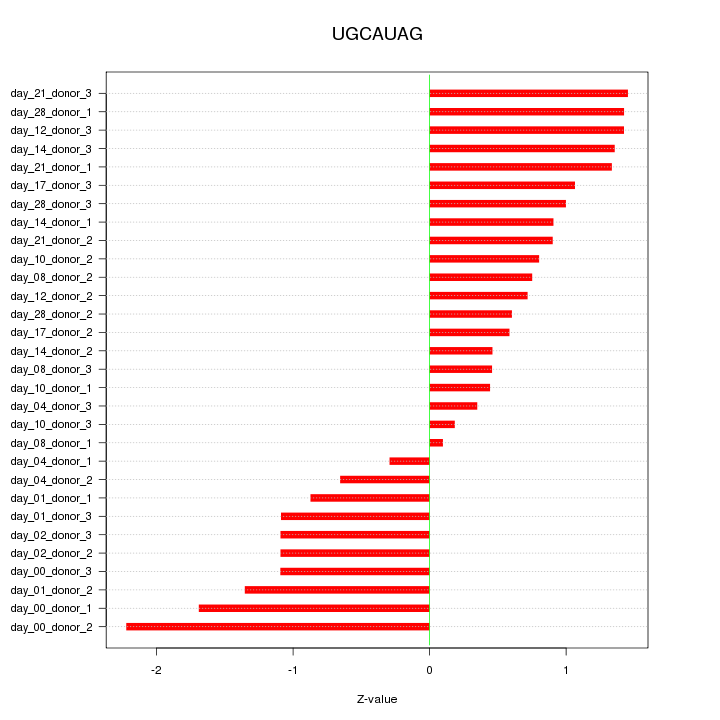
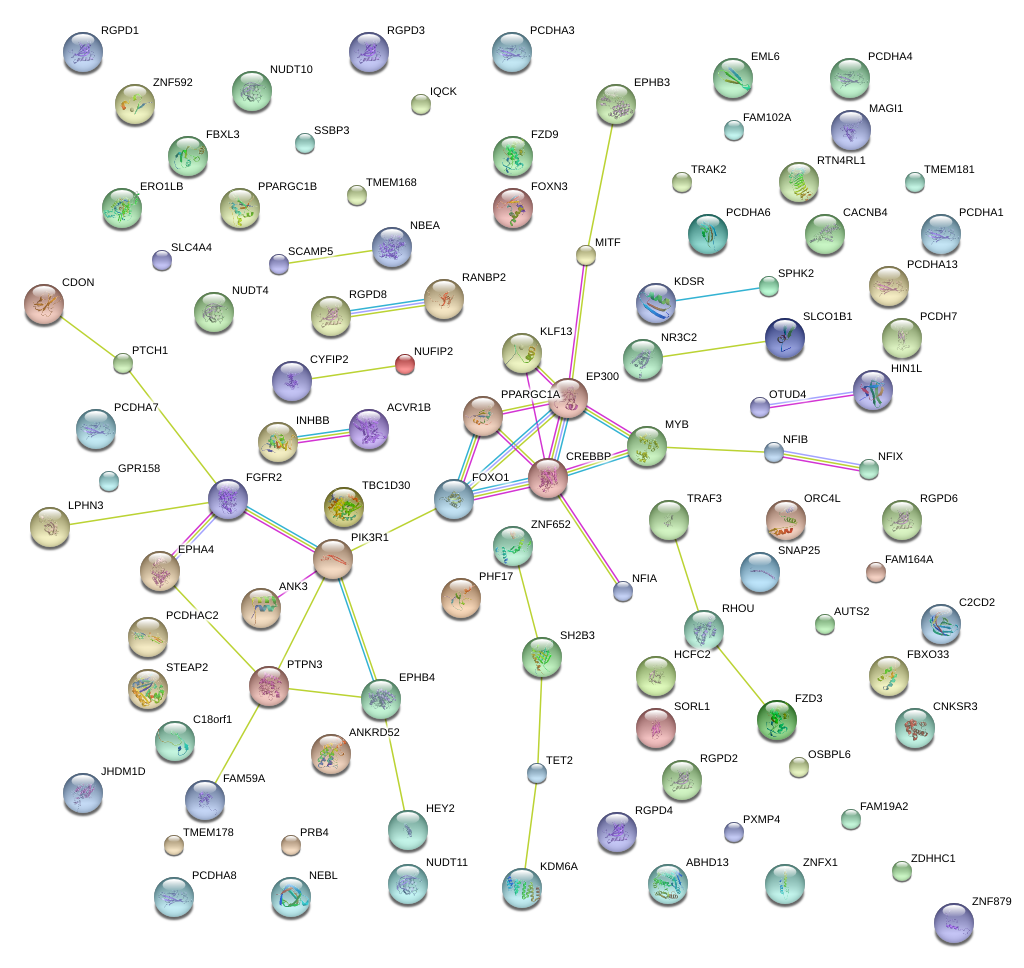
|
chr5_+_156693051
|
5.278
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr5_+_156696351
|
5.062
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr4_-_149363498
|
4.511
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr13_+_36050885
|
4.344
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr2_+_121103670
|
4.163
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr4_+_72052354
|
4.014
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr1_+_228870801
|
3.988
|
NM_021205
|
RHOU
|
ras homolog gene family, member U
|
|
chr13_+_35516390
|
3.749
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr1_+_61547625
|
2.917
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_61542928
|
2.770
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr2_+_39892637
|
2.685
|
NM_001167959
|
TMEM178
|
transmembrane protein 178
|
|
chr1_-_236445294
|
2.596
|
NM_019891
|
ERO1LB
|
ERO1-like beta (S. cerevisiae)
|
|
chr5_+_140186658
|
2.542
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr5_+_140254930
|
2.502
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr2_+_39893021
|
2.499
|
NM_152390
|
TMEM178
|
transmembrane protein 178
|
|
chr5_+_140213968
|
2.484
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr5_+_140220811
|
2.448
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr1_+_61547388
|
2.267
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr15_+_75287829
|
2.192
|
NM_001178111
NM_001178112
NM_138967
|
SCAMP5
|
secretory carrier membrane protein 5
|
|
chr4_+_72204769
|
2.178
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr2_-_222436996
|
2.096
|
NM_004438
|
EPHA4
|
EPH receptor A4
|
|
chr2_-_202316227
|
2.095
|
NM_015049
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr5_+_140247767
|
2.038
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr9_-_130742803
|
1.902
|
NM_001035254
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr3_+_69812961
|
1.805
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr4_-_23891608
|
1.798
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr6_+_126070724
|
1.771
|
NM_012259
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr20_+_54933982
|
1.674
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr12_+_65218351
|
1.639
|
NM_015279
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr5_+_140165875
|
1.590
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr16_-_67450149
|
1.575
|
NM_013304
|
ZDHHC1
|
zinc finger, DHHC-type containing 1
|
|
chr3_+_69788562
|
1.553
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_+_69812620
|
1.552
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr19_+_49122789
|
1.547
|
NM_001204159
|
SPHK2
|
sphingosine kinase 2
|
|
chr3_+_69915374
|
1.541
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr5_+_140207562
|
1.540
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr6_+_135502445
|
1.534
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr21_-_43373938
|
1.525
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr12_+_104458235
|
1.481
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr14_-_90085325
|
1.476
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr19_+_49122547
|
1.474
|
NM_001204158
NM_001243876
NM_020126
|
SPHK2
|
sphingosine kinase 2
|
|
chr14_-_89883306
|
1.437
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr5_+_149109814
|
1.435
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr2_+_54952148
|
1.431
|
NM_001039753
|
EML6
|
echinoderm microtubule associated protein like 6
|
|
chr10_+_25464289
|
1.422
|
NM_020752
|
GPR158
|
G protein-coupled receptor 158
|
|
chr19_+_49128208
|
1.412
|
NM_001204160
|
SPHK2
|
sphingosine kinase 2
|
|
chr14_-_39901619
|
1.405
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr7_+_89840999
|
1.404
|
NM_001244944
NM_001244945
NM_152999
NM_001040665
NM_001244946
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr9_-_130712866
|
1.385
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr5_+_149151502
|
1.367
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr4_+_30721865
|
1.325
|
NM_032456
NM_001173523
NM_002589
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr17_-_47439326
|
1.321
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr3_-_161089747
|
1.313
|
NM_001040100
|
SPTSSB
|
serine palmitoyltransferase, small subunit B
|
|
chr10_-_62149487
|
1.299
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr18_-_30050394
|
1.297
|
NM_001242409
NM_022751
|
FAM59A
|
family with sequence similarity 59, member A
|
|
chr10_-_61900659
|
1.268
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr20_-_47894591
|
1.268
|
NM_021035
|
ZNFX1
|
zinc finger, NFX1-type containing 1
|
|
chr5_+_140345746
|
1.262
|
NM_018899
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr5_+_140180782
|
1.220
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr10_-_62493282
|
1.217
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr7_-_112430408
|
1.206
|
NM_022484
|
TMEM168
|
transmembrane protein 168
|
|
chr10_-_62332666
|
1.201
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr10_-_21463019
|
1.187
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr11_-_125933186
|
1.179
|
NM_001243597
NM_016952
|
CDON
|
Cdon homolog (mouse)
|
|
chr2_-_148779172
|
1.174
|
NM_181742
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr12_+_93771607
|
1.152
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr14_+_103243813
|
1.150
|
NM_001199427
NM_003300
NM_145725
NM_145726
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr8_+_79578258
|
1.129
|
NM_016010
|
FAM164A
|
family with sequence similarity 164, member A
|
|
chr5_+_140235594
|
1.107
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr2_+_179059071
|
1.093
|
NM_001201480
NM_001201481
NM_001201482
NM_032523
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr10_-_123356158
|
1.080
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr5_+_140261792
|
1.070
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr16_-_3930120
|
1.065
|
NM_001079846
NM_004380
|
CREBBP
|
CREB binding protein
|
|
chr11_+_121322848
|
1.063
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr12_-_56652118
|
1.059
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr9_-_98270321
|
1.020
|
NM_001083604
NM_001083605
|
PTCH1
|
patched 1
|
|
chr17_-_1928177
|
1.011
|
NM_178568
|
RTN4RL1
|
reticulon 4 receptor-like 1
|
|
chr8_+_28351694
|
1.006
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr12_+_52345437
|
0.973
|
NM_004302
NM_020328
|
ACVR1B
|
activin A receptor, type IB
|
|
chrX_+_44732402
|
0.968
|
NM_021140
|
KDM6A
|
lysine (K)-specific demethylase 6A
|
|
chr15_+_31619043
|
0.962
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr12_+_111843743
|
0.959
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr9_-_98269686
|
0.959
|
NM_001083606
|
PTCH1
|
patched 1
|
|
chr9_-_112260529
|
0.940
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr10_-_123353771
|
0.935
|
NM_001144916
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr20_+_10199455
|
0.928
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr13_+_108870713
|
0.917
|
NM_032859
|
ABHD13
|
abhydrolase domain containing 13
|
|
chr13_-_77601296
|
0.911
|
NM_012158
|
FBXL3
|
F-box and leucine-rich repeat protein 3
|
|
chr3_+_69985750
|
0.909
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr5_+_67511578
|
0.896
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr16_+_19727698
|
0.874
|
NM_153208
|
IQCK
|
IQ motif containing K
|
|
chr2_-_148778262
|
0.871
|
NM_001190882
NM_001190879
NM_002552
NM_001190881
NM_181741
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr7_+_69063768
|
0.868
|
NM_001127231
NM_001127232
NM_015570
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr18_+_13218777
|
0.867
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr3_-_66024508
|
0.864
|
NM_001033057
NM_004742
NM_015520
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr20_-_32307889
|
0.839
|
NM_007238
NM_183397
|
PXMP4
|
peroxisomal membrane protein 4, 24kDa
|
|
chr2_-_152955502
|
0.828
|
NM_000726
NM_001145798
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr15_+_85291802
|
0.827
|
NM_014630
|
ZNF592
|
zinc finger protein 592
|
|
chr9_-_98269480
|
0.826
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr9_-_98270830
|
0.820
|
NM_000264
|
PTCH1
|
patched 1
|
|
chr5_+_178450775
|
0.814
|
NM_001136116
|
ZNF879
|
zinc finger protein 879
|
|
chr7_-_139876718
|
0.811
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr17_-_27621163
|
0.810
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr4_-_146100793
|
0.798
|
NM_001102653
|
OTUD4
|
OTU domain containing 4
|
|
chr13_-_41240607
|
0.795
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr18_-_61034350
|
0.794
|
NM_002035
|
KDSR
|
3-ketodihydrosphingosine reductase
|
|
chr2_-_152955208
|
0.787
|
NM_001005746
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr4_+_106067754
|
0.786
|
NM_001127208
|
TET2
|
tet methylcytosine dioxygenase 2
|
|
chr4_+_129730772
|
0.784
|
NM_024900
NM_199320
|
PHF17
|
PHD finger protein 17
|
|
chr10_-_21186530
|
0.779
|
NM_006393
|
NEBL
|
nebulette
|
|
chr1_-_54871978
|
0.775
|
NM_018070
NM_145716
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr6_+_158957467
|
0.774
|
NM_020823
|
TMEM181
|
transmembrane protein 181
|
|
chr2_+_109335906
|
0.774
|
NM_006267
|
RANBP2
|
RAN binding protein 2
|
|
chr11_-_73309090
|
0.772
|
NM_015159
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr11_+_118307072
|
0.758
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr1_-_54872075
|
0.753
|
NM_001009955
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr5_+_112312392
|
0.737
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr2_+_179345194
|
0.730
|
NM_019091
|
PLEKHA3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3
|
|
chrX_+_10124984
|
0.718
|
NM_001830
|
CLCN4
|
chloride channel 4
|
|
chr4_+_108745718
|
0.716
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr1_+_95558072
|
0.711
|
NM_001199679
|
TMEM56
|
transmembrane protein 56
|
|
chr18_-_53255734
|
0.706
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr3_-_52931595
|
0.706
|
NM_001198974
NM_198563
|
TMEM110-MUSTN1
TMEM110
|
TMEM110-MUSTN1 readthrough
transmembrane protein 110
|
|
chr17_-_56065596
|
0.703
|
NM_007146
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chr18_+_13620866
|
0.697
|
NM_004338
NM_181483
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr4_+_108814419
|
0.694
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr13_-_110438896
|
0.692
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr8_-_103876390
|
0.686
|
NM_015878
NM_148174
|
AZIN1
|
antizyme inhibitor 1
|
|
chr18_+_13611664
|
0.684
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr6_-_139695349
|
0.682
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr4_-_170924911
|
0.679
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr6_-_79787939
|
0.679
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr16_+_68119211
|
0.670
|
NM_004555
NM_173163
NM_173165
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3
|
|
chr4_+_26862312
|
0.667
|
NM_001169117
NM_001169118
NM_020860
|
STIM2
|
stromal interaction molecule 2
|
|
chr7_+_43152196
|
0.667
|
NM_015052
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1
|
|
chr1_-_205091137
|
0.662
|
NM_001193272
NM_001193273
NM_005057
|
RBBP5
|
retinoblastoma binding protein 5
|
|
chr5_+_67584251
|
0.662
|
NM_181524
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr9_-_98279246
|
0.662
|
NM_001083602
NM_001083603
|
PTCH1
|
patched 1
|
|
chr4_+_57775064
|
0.660
|
NM_001193508
|
REST
|
RE1-silencing transcription factor
|
|
chr1_-_204329043
|
0.659
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr2_-_174828775
|
0.655
|
NM_001017371
|
SP3
|
Sp3 transcription factor
|
|
chr20_-_31071187
|
0.651
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr1_-_39338976
|
0.650
|
NM_012333
|
MYCBP
|
c-myc binding protein
|
|
chr16_+_27561441
|
0.649
|
NM_015202
|
KIAA0556
|
KIAA0556
|
|
chr14_-_61190458
|
0.649
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr6_+_144612872
|
0.646
|
NM_007124
|
UTRN
|
utrophin
|
|
chr6_-_139695784
|
0.644
|
NM_006079
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr8_-_95961543
|
0.638
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr2_+_148602569
|
0.637
|
NM_001616
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr22_-_31742217
|
0.636
|
NM_014323
NM_032050
NM_032051
NM_032052
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr15_+_69452799
|
0.629
|
NM_015554
|
GLCE
|
glucuronic acid epimerase
|
|
chr21_-_38338618
|
0.621
|
NM_001242784
|
HLCS
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase)
|
|
chr20_+_33292117
|
0.620
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr10_+_45455149
|
0.618
|
NM_032023
|
RASSF4
|
Ras association (RalGDS/AF-6) domain family member 4
|
|
chr2_-_152830448
|
0.613
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr1_-_40367539
|
0.608
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr5_+_139493689
|
0.604
|
NM_005859
|
PURA
|
purine-rich element binding protein A
|
|
chr5_-_133968463
|
0.603
|
NM_001033503
NM_016103
|
SAR1B
|
SAR1 homolog B (S. cerevisiae)
|
|
chr7_+_37960162
|
0.602
|
NM_001242946
NM_017549
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr1_+_95582827
|
0.599
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr16_-_87525317
|
0.596
|
NM_015144
|
ZCCHC14
|
zinc finger, CCHC domain containing 14
|
|
chr4_+_144434555
|
0.591
|
NM_003601
|
SMARCA5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5
|
|
chr1_+_167189948
|
0.590
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr21_-_38362481
|
0.587
|
NM_000411
|
HLCS
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase)
|
|
chr11_+_109964086
|
0.587
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr21_-_43346780
|
0.578
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr2_-_24149899
|
0.565
|
NM_001242338
NM_017552
|
ATAD2B
|
ATPase family, AAA domain containing 2B
|
|
chr17_-_4046244
|
0.564
|
NM_015113
|
ZZEF1
|
zinc finger, ZZ-type with EF-hand domain 1
|
|
chr13_-_111567415
|
0.562
|
NM_017664
|
ANKRD10
|
ankyrin repeat domain 10
|
|
chrX_-_48828250
|
0.560
|
NM_004979
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1
|
|
chr2_+_179184970
|
0.550
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr10_-_123357503
|
0.547
|
NM_000141
NM_001144917
NM_001144918
NM_001144919
NM_022970
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr10_+_129845806
|
0.542
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr8_+_67687415
|
0.542
|
NM_013257
NM_170709
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr8_-_70745624
|
0.536
|
NM_001146008
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr1_+_100598705
|
0.533
|
NM_019083
|
CCDC76
|
coiled-coil domain containing 76
|
|
chr4_-_125633716
|
0.533
|
NM_001167882
NM_020337
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr22_+_40440664
|
0.528
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chrX_+_40944858
|
0.527
|
NM_001039590
NM_001039591
|
USP9X
|
ubiquitin specific peptidase 9, X-linked
|
|
chr15_-_52970808
|
0.527
|
NM_019600
|
FAM214A
|
family with sequence similarity 214, member A
|
|
chr5_+_67586466
|
0.522
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr1_+_218518675
|
0.515
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr1_+_167298280
|
0.513
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr16_+_71879893
|
0.512
|
NM_001137675
|
ATXN1L
|
ataxin 1-like
|
|
chr8_-_91657898
|
0.510
|
NM_001008495
NM_001146273
|
TMEM64
|
transmembrane protein 64
|
|
chr5_-_88199868
|
0.506
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr5_+_140787769
|
0.503
|
NM_018926
NM_032100
|
PCDHGB6
|
protocadherin gamma subfamily B, 6
|
|
chr17_+_58677489
|
0.497
|
NM_003620
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr6_+_170102236
|
0.492
|
NM_001029863
|
C6orf120
|
chromosome 6 open reading frame 120
|
|
chr8_+_67624652
|
0.492
|
NM_001033578
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr3_-_113415313
|
0.489
|
NM_001009899
|
KIAA2018
|
KIAA2018
|
|
chr1_+_36348795
|
0.481
|
NM_012199
|
EIF2C1
|
eukaryotic translation initiation factor 2C, 1
|
|
chr14_+_58765209
|
0.479
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr8_+_98881277
|
0.479
|
NM_002380
NM_030583
|
MATN2
|
matrilin 2
|
|
chr5_-_43557520
|
0.478
|
NM_183323
|
PAIP1
|
poly(A) binding protein interacting protein 1
|
|
chr10_-_133109983
|
0.477
|
NM_174937
|
TCERG1L
|
transcription elongation regulator 1-like
|
|
chr1_+_41249479
|
0.468
|
NM_004700
NM_172163
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr10_-_94333847
|
0.466
|
NM_004969
|
IDE
|
insulin-degrading enzyme
|
|
chr17_+_65821625
|
0.463
|
NM_004459
NM_182641
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr9_-_112213663
|
0.462
|
NM_001145369
NM_001145370
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|