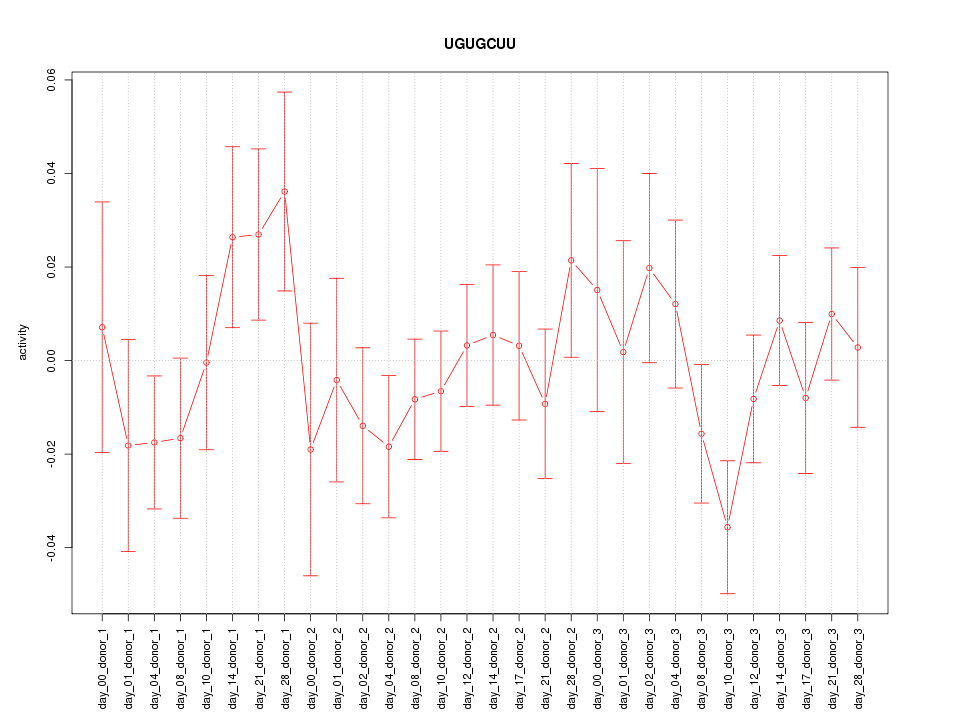
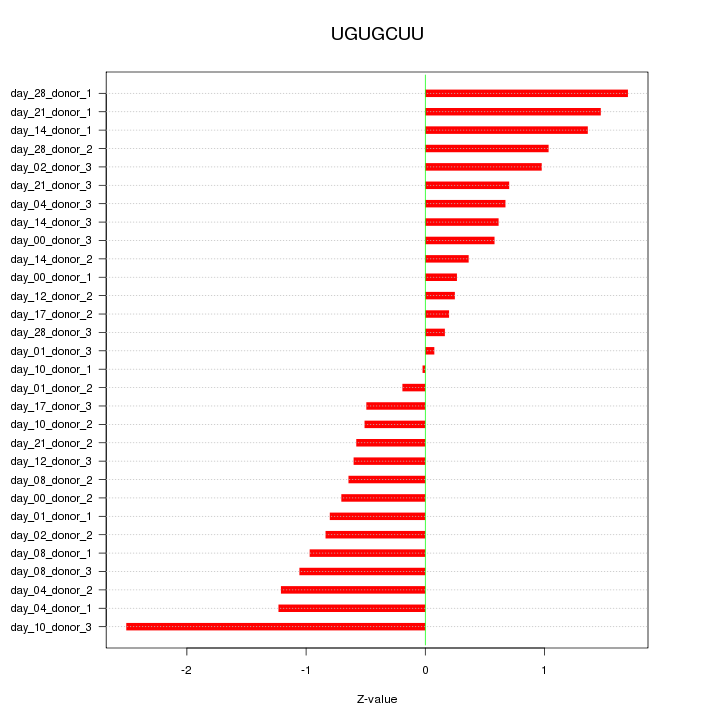
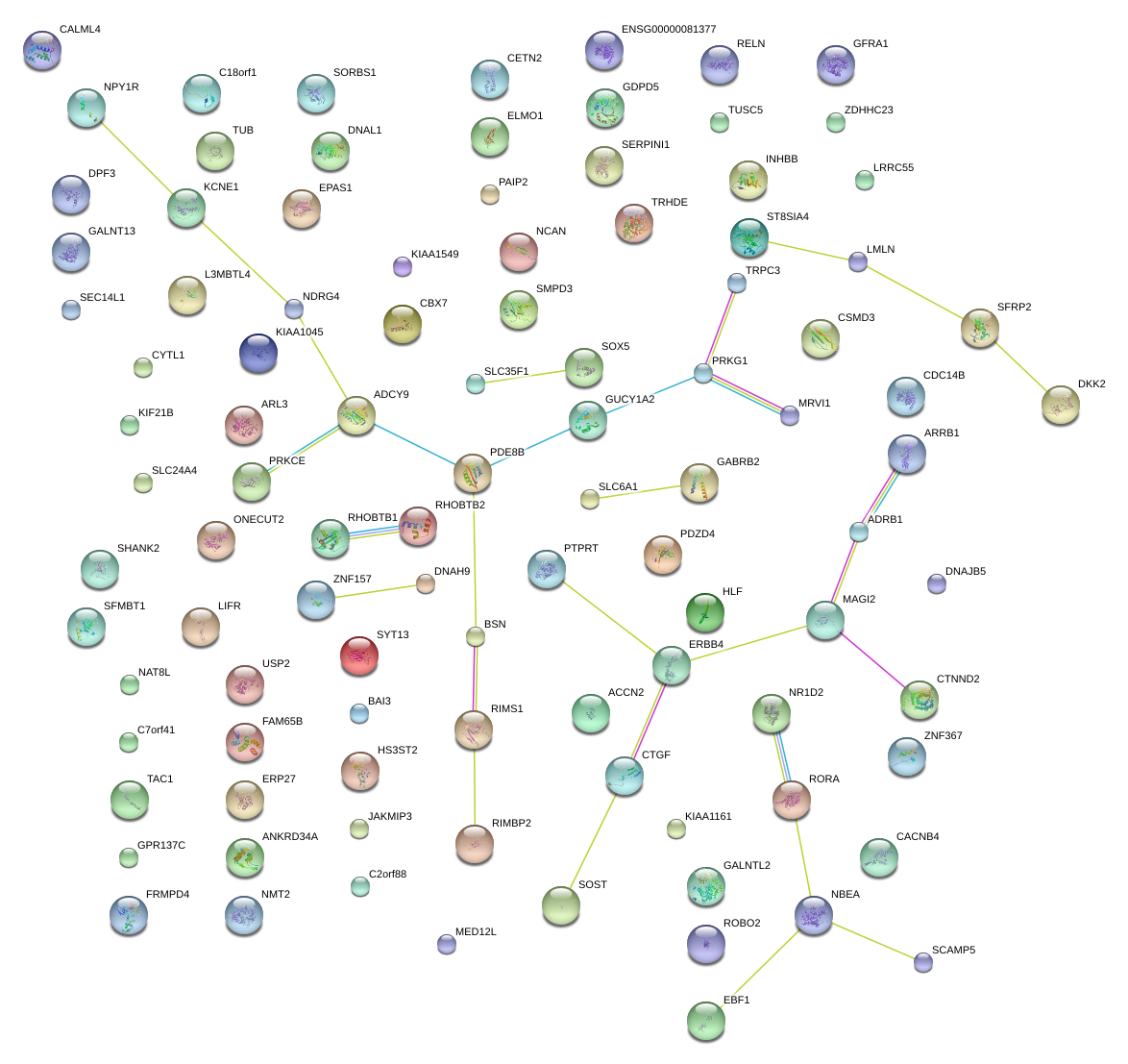
|
chr6_-_24911194
|
1.595
|
NM_014722
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr6_-_132272311
|
1.414
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr7_-_37024664
|
1.382
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr7_-_37393271
|
1.339
|
NM_001206482
|
ELMO1
|
engulfment and cell motility 1
|
|
chr7_-_37488554
|
1.336
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr7_-_37488894
|
1.283
|
NM_001206480
|
ELMO1
|
engulfment and cell motility 1
|
|
chr4_-_154710207
|
1.133
|
NM_003013
|
SFRP2
|
secreted frizzled-related protein 2
|
|
chr6_+_69344939
|
1.122
|
NM_001704
|
BAI3
|
brain-specific angiogenesis inhibitor 3
|
|
chr20_-_41818464
|
1.105
|
NM_007050
NM_133170
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chr2_-_213403280
|
1.008
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr12_-_131002409
|
0.935
|
NM_015347
|
RIMBP2
|
RIMS binding protein 2
|
|
chr11_+_8102691
|
0.931
|
NM_177972
|
TUB
|
tubby homolog (mouse)
|
|
chr18_-_6414900
|
0.868
|
NM_173464
|
L3MBTL4
|
l(3)mbt-like 4 (Drosophila)
|
|
chr1_-_200992824
|
0.867
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr14_+_92790151
|
0.831
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr14_+_92789536
|
0.818
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr18_+_13218777
|
0.810
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr2_+_191014971
|
0.795
|
NM_001042520
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr14_+_53019865
|
0.790
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr17_+_53342320
|
0.767
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr12_+_50451486
|
0.761
|
NM_001095
NM_020039
|
ACCN2
|
amiloride-sensitive cation channel 2, neuronal
|
|
chr18_+_13620866
|
0.748
|
NM_004338
NM_181483
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr2_+_191002485
|
0.726
|
NM_001042521
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr18_+_13611664
|
0.720
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr10_-_62703822
|
0.717
|
NM_001242359
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr5_-_38556682
|
0.714
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr10_+_133918312
|
0.712
|
NM_001105521
|
JAKMIP3
|
Janus kinase and microtubule interacting protein 3
|
|
chr14_+_92788924
|
0.712
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr13_+_36050885
|
0.703
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr15_-_60884615
|
0.700
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr4_-_107957452
|
0.692
|
NM_014421
|
DKK2
|
dickkopf 2 homolog (Xenopus laevis)
|
|
chr15_-_68498373
|
0.689
|
NM_001031733
NM_033429
|
CALML4
|
calmodulin-like 4
|
|
chr2_+_191045588
|
0.687
|
NM_001042519
NM_032321
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr19_+_19322763
|
0.674
|
NM_004386
|
NCAN
|
neurocan
|
|
chr14_-_73360792
|
0.667
|
NM_012074
|
DPF3
|
D4, zinc and double PHD fingers, family 3
|
|
chr2_+_121103670
|
0.659
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr13_+_35516390
|
0.657
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr3_+_113666544
|
0.654
|
NM_173570
|
ZDHHC23
|
zinc finger, DHHC-type containing 23
|
|
chr5_-_38595506
|
0.648
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr11_-_106888829
|
0.640
|
NM_000855
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2
|
|
chr5_-_100238870
|
0.631
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr11_-_75236418
|
0.630
|
NM_030792
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5
|
|
chr3_+_197687070
|
0.626
|
NM_001136049
NM_033029
|
LMLN
|
leishmanolysin-like (metallopeptidase M8 family)
|
|
chr11_-_70507911
|
0.616
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr6_+_72926462
|
0.615
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr15_-_61521478
|
0.611
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr22_-_39548448
|
0.608
|
NM_175709
|
CBX7
|
chromobox homolog 7
|
|
chr11_-_119234859
|
0.597
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr7_+_30174343
|
0.595
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr16_+_22825806
|
0.589
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr10_-_118031542
|
0.585
|
NM_145793
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr10_-_118032975
|
0.578
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr2_-_152955502
|
0.568
|
NM_000726
NM_001145798
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr10_-_118032717
|
0.564
|
NM_001145453
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr3_+_11034419
|
0.562
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr3_+_49591897
|
0.559
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr5_+_76506705
|
0.559
|
NM_001029851
NM_001029852
NM_001029853
NM_001029854
NM_003719
|
PDE8B
|
phosphodiesterase 8B
|
|
chr7_-_79082870
|
0.553
|
NM_012301
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr10_-_97321094
|
0.545
|
NM_015385
NM_024991
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr6_+_72922513
|
0.537
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr7_-_103629962
|
0.534
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr2_-_152955208
|
0.518
|
NM_001005746
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr8_-_114389381
|
0.516
|
NM_198124
|
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chr3_+_167453431
|
0.513
|
NM_001122752
NM_005025
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1
|
|
chr16_-_68482373
|
0.511
|
NM_018667
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II)
|
|
chr11_-_45307617
|
0.510
|
NM_001247987
NM_020826
|
SYT13
|
synaptotagmin XIII
|
|
chr10_+_115803805
|
0.503
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr11_-_70935807
|
0.499
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr17_+_75084724
|
0.490
|
NM_001204408
|
SEC14L1
LINC00338
|
SEC14-like 1 (S. cerevisiae)
long intergenic non-protein coding RNA 338
|
|
chr14_+_74111577
|
0.490
|
NM_001201366
NM_031427
|
DNAL1
|
dynein, axonemal, light chain 1
|
|
chr5_-_160975124
|
0.484
|
NM_000813
NM_021911
|
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2
|
|
chr17_+_1182956
|
0.482
|
NM_172367
|
TUSC5
|
tumor suppressor candidate 5
|
|
chr9_-_34376831
|
0.481
|
NM_020702
|
KIAA1161
|
KIAA1161
|
|
chr6_+_72926144
|
0.479
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr15_-_60919642
|
0.462
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr7_+_97361270
|
0.460
|
NM_003182
NM_013996
NM_013997
NM_013998
|
TAC1
|
tachykinin, precursor 1
|
|
chr7_-_138665853
|
0.454
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr15_+_75287829
|
0.450
|
NM_001178111
NM_001178112
NM_138967
|
SCAMP5
|
secretory carrier membrane protein 5
|
|
chr11_-_10715503
|
0.447
|
NM_001100163
NM_001206880
NM_001206881
NM_130385
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr3_-_53080038
|
0.444
|
NM_001005158
NM_016329
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr16_+_19535178
|
0.441
|
NM_001199022
NM_014711
|
CCP110
|
centriolar coiled coil protein 110kDa
|
|
chr3_-_53079285
|
0.432
|
NM_001005159
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr17_+_75085234
|
0.430
|
NM_001204410
|
SEC14L1
LINC00338
|
SEC14-like 1 (S. cerevisiae)
long intergenic non-protein coding RNA 338
|
|
chr21_-_35831856
|
0.423
|
NM_001127668
NM_001127669
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr17_+_75136985
|
0.421
|
NM_001039573
NM_001143998
NM_001143999
NM_003003
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae)
|
|
chr2_-_152830448
|
0.414
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr21_-_35883570
|
0.414
|
NM_000219
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr6_+_118228688
|
0.410
|
NM_001029858
|
SLC35F1
|
solute carrier family 35, member F1
|
|
chrX_+_12156584
|
0.406
|
NM_014728
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr16_+_58534045
|
0.400
|
NM_001242833
NM_001242834
NM_001242835
NM_001242836
|
NDRG4
|
NDRG family member 4
|
|
chr1_+_145470507
|
0.399
|
NM_001039888
|
ANKRD34A
|
ankyrin repeat domain 34A
|
|
chr6_+_73076431
|
0.397
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr5_-_158526698
|
0.395
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr4_-_122854192
|
0.395
|
NM_003305
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3
|
|
chr9_+_34958171
|
0.393
|
NM_015297
|
KIAA1045
|
KIAA1045
|
|
chr21_-_35828038
|
0.385
|
NM_001127670
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr11_-_119252364
|
0.382
|
NM_001243759
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr3_+_77089248
|
0.380
|
NM_002942
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila)
|
|
chr3_+_23987514
|
0.379
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr8_-_114449018
|
0.379
|
NM_052900
NM_198123
|
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chr2_+_154728424
|
0.375
|
NM_052917
|
GALNT13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13)
|
|
chr9_-_99382073
|
0.373
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr18_+_55102777
|
0.373
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr5_-_11903964
|
0.369
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr5_+_138678130
|
0.364
|
NM_001033112
|
PAIP2
|
poly(A) binding protein interacting protein 2
|
|
chr12_-_15091441
|
0.363
|
NM_152321
|
ERP27
|
endoplasmic reticulum protein 27
|
|
chr10_-_15210610
|
0.361
|
NM_004808
|
NMT2
|
N-myristoyltransferase 2
|
|
chrX_-_151999259
|
0.361
|
NM_004344
|
CETN2
|
centrin, EF-hand protein, 2
|
|
chr16_-_4166185
|
0.361
|
NM_001116
|
ADCY9
|
adenylate cyclase 9
|
|
chr3_+_150804675
|
0.360
|
NM_053002
|
MED12L
|
mediator complex subunit 12-like
|
|
chr11_+_56949220
|
0.360
|
NM_001005210
|
LRRC55
|
leucine rich repeat containing 55
|
|
chr16_+_58497993
|
0.360
|
NM_001130487
|
NDRG4
|
NDRG family member 4
|
|
chr6_+_72596648
|
0.356
|
NM_014989
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr4_-_164253946
|
0.356
|
NM_000909
|
NPY1R
|
neuropeptide Y receptor Y1
|
|
chr16_+_58497548
|
0.356
|
NM_020465
|
NDRG4
|
NDRG family member 4
|
|
chr17_-_41836132
|
0.356
|
NM_025237
|
SOST
|
sclerostin
|
|
chr3_+_23986735
|
0.355
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr2_+_45878912
|
0.353
|
NM_005400
|
PRKCE
|
protein kinase C, epsilon
|
|
chr12_+_72666462
|
0.352
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr12_-_23737507
|
0.349
|
NM_178010
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr10_-_97200895
|
0.342
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr10_+_52834233
|
0.342
|
NM_006258
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr11_-_75062650
|
0.341
|
NM_004041
NM_020251
|
ARRB1
|
arrestin, beta 1
|
|
chr4_+_2061238
|
0.341
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr10_-_104473957
|
0.339
|
NM_004311
|
ARL3
|
ADP-ribosylation factor-like 3
|
|
chrX_-_153095996
|
0.339
|
NM_032512
|
PDZD4
|
PDZ domain containing 4
|
|
chr1_-_119532178
|
0.337
|
NM_152380
|
TBX15
|
T-box 15
|
|
chr12_+_26348488
|
0.334
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr17_-_46507481
|
0.332
|
NM_001075099
NM_003726
|
SKAP1
|
src kinase associated phosphoprotein 1
|
|
chr6_+_73331570
|
0.332
|
NM_001160130
NM_001160132
NM_001160133
NM_001160134
NM_019842
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr19_-_55954229
|
0.332
|
NM_001145176
|
SHISA7
|
shisa homolog 7 (Xenopus laevis)
|
|
chr12_-_24715349
|
0.330
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr8_-_15095791
|
0.328
|
NM_139167
|
SGCZ
|
sarcoglycan, zeta
|
|
chr8_-_70745624
|
0.328
|
NM_001146008
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr20_+_43990576
|
0.327
|
NM_001197129
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr7_+_138916230
|
0.326
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr11_-_86666211
|
0.323
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr16_+_58498725
|
0.321
|
NM_022910
|
NDRG4
|
NDRG family member 4
|
|
chr4_-_122872908
|
0.321
|
NM_001130698
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3
|
|
chr11_+_93517394
|
0.321
|
NM_004268
|
MED17
|
mediator complex subunit 17
|
|
chr8_-_70745403
|
0.320
|
NM_001146009
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr5_+_139493689
|
0.317
|
NM_005859
|
PURA
|
purine-rich element binding protein A
|
|
chr11_-_10673832
|
0.313
|
NM_001098579
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr5_-_131347518
|
0.311
|
NM_001205248
NM_001205251
NM_001205250
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr16_-_52580805
|
0.311
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr5_-_131347249
|
0.309
|
NM_001009185
NM_001205247
NM_015256
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr20_-_4795768
|
0.307
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr18_+_67956136
|
0.307
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr15_+_59730272
|
0.307
|
NM_152450
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chr12_+_26348268
|
0.306
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr9_+_130374466
|
0.305
|
NM_001032221
NM_003165
|
STXBP1
|
syntaxin binding protein 1
|
|
chr20_-_4804067
|
0.300
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr3_-_179754516
|
0.298
|
NM_016559
|
PEX5L
|
peroxisomal biogenesis factor 5-like
|
|
chr19_-_38720312
|
0.297
|
NM_001135156
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr13_-_79233266
|
0.295
|
NM_024546
|
RNF219
|
ring finger protein 219
|
|
chr17_+_75181294
|
0.294
|
NM_001144001
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae)
|
|
chr4_+_85504056
|
0.293
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr15_-_27018216
|
0.292
|
NM_000814
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr3_+_16926451
|
0.291
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr19_-_38714781
|
0.290
|
NM_001135155
NM_004647
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr1_+_110693131
|
0.286
|
NM_001010898
|
SLC6A17
|
solute carrier family 6, member 17
|
|
chr20_+_51588945
|
0.283
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr15_+_74422589
|
0.281
|
NM_001130138
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr12_+_78225068
|
0.279
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr16_-_62070738
|
0.278
|
NM_001796
|
CDH8
|
cadherin 8, type 2
|
|
chr7_-_71802207
|
0.278
|
NM_001017440
|
CALN1
|
calneuron 1
|
|
chr6_+_148663728
|
0.276
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr3_+_238274
|
0.271
|
NM_001253387
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr10_+_43572500
|
0.271
|
NM_020630
NM_020975
|
RET
|
ret proto-oncogene
|
|
chr3_-_114343052
|
0.271
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr7_-_82792127
|
0.270
|
NM_014510
NM_033026
|
PCLO
|
piccolo (presynaptic cytomatrix protein)
|
|
chr3_+_33318911
|
0.270
|
NM_012157
NM_001171713
|
FBXL2
|
F-box and leucine-rich repeat protein 2
|
|
chr3_-_46037299
|
0.266
|
NM_024513
|
FYCO1
|
FYVE and coiled-coil domain containing 1
|
|
chr7_-_102789334
|
0.265
|
NM_001122838
NM_198990
|
NAPEPLD
|
N-acyl phosphatidylethanolamine phospholipase D
|
|
chr10_+_119000583
|
0.265
|
NM_003054
|
SLC18A2
|
solute carrier family 18 (vesicular monoamine), member 2
|
|
chr8_-_70747200
|
0.264
|
NM_030958
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr5_+_155753755
|
0.263
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chrX_-_129402753
|
0.263
|
NM_017666
|
ZNF280C
|
zinc finger protein 280C
|
|
chr12_-_71182699
|
0.262
|
NM_001207015
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr7_+_5253816
|
0.258
|
NM_001033520
|
WIPI2
|
WD repeat domain, phosphoinositide interacting 2
|
|
chr3_-_114102488
|
0.258
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr2_-_217236666
|
0.256
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr4_+_129730772
|
0.256
|
NM_024900
NM_199320
|
PHF17
|
PHD finger protein 17
|
|
chrX_-_117251302
|
0.255
|
NM_001168301
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr18_-_11148760
|
0.254
|
NM_022068
|
PIEZO2
|
piezo-type mechanosensitive ion channel component 2
|
|
chr7_+_114055048
|
0.254
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr8_-_41754244
|
0.253
|
NM_001142446
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr3_-_114343791
|
0.251
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr15_+_74421698
|
0.247
|
NM_001130136
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr5_-_39074494
|
0.245
|
NM_152756
|
RICTOR
|
RPTOR independent companion of MTOR, complex 2
|
|
chrX_+_64708614
|
0.244
|
NM_001010888
|
ZC3H12B
|
zinc finger CCCH-type containing 12B
|
|
chr3_+_187871473
|
0.244
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chrX_-_117107700
|
0.243
|
NM_033495
NM_001168300
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr12_-_56652118
|
0.241
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr1_+_61547625
|
0.241
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chrX_-_117250624
|
0.240
|
NM_001168302
NM_001168303
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr6_-_94129059
|
0.239
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr3_-_66550844
|
0.237
|
NM_015541
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr10_+_22610138
|
0.234
|
NM_005180
|
BMI1
|
BMI1 polycomb ring finger oncogene
|
|
chr15_+_59063389
|
0.234
|
NM_001040450
NM_001040453
|
FAM63B
|
family with sequence similarity 63, member B
|