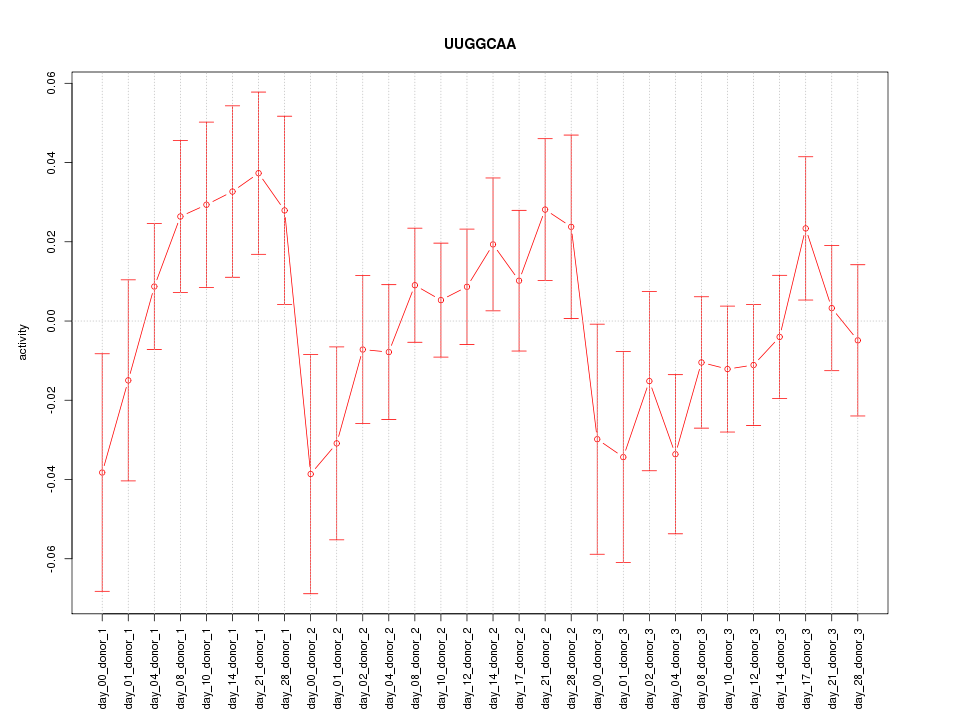
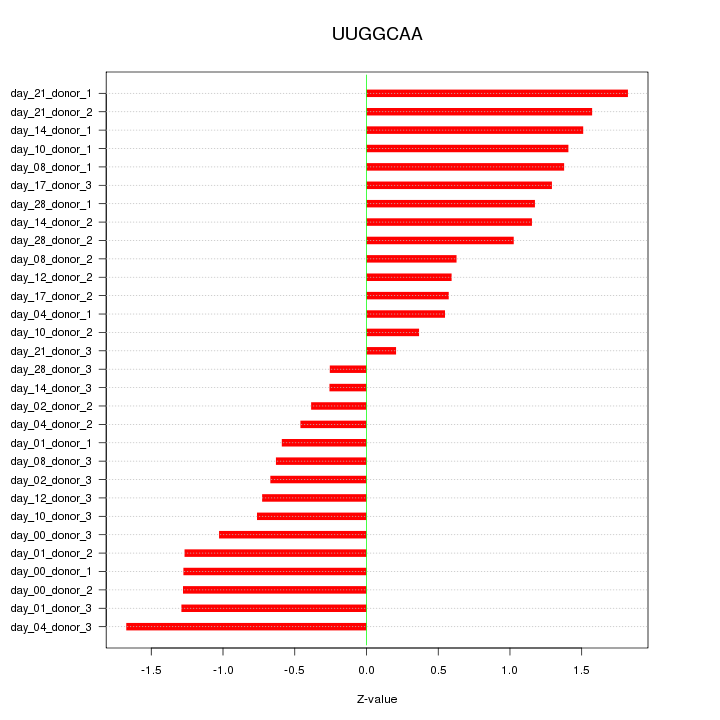
|
chr16_-_52580805
|
6.517
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr16_-_52581713
|
6.225
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr7_-_131241321
|
4.166
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr2_-_99771186
|
4.101
|
NM_025244
|
TSGA10
|
testis specific, 10
|
|
chr20_+_9049660
|
4.076
|
NM_001172646
|
PLCB4
|
phospholipase C, beta 4
|
|
chr2_-_99758034
|
4.040
|
NM_182911
|
TSGA10
|
testis specific, 10
|
|
chr6_-_10838712
|
3.892
|
NM_001242385
NM_001242957
NM_005906
|
MAK
|
male germ cell-associated kinase
|
|
chr20_+_9288446
|
3.841
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr7_-_158380360
|
3.726
|
NM_002847
NM_130842
NM_130843
|
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr6_+_135502445
|
3.693
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr5_-_16509106
|
3.636
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr20_+_9198036
|
3.591
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr13_+_24734788
|
3.381
|
NM_001166271
NM_153023
|
SPATA13
|
spermatogenesis associated 13
|
|
chr11_+_111848008
|
3.305
|
NM_033425
|
DIXDC1
|
DIX domain containing 1
|
|
chr3_-_9994057
|
3.224
|
NM_207351
|
PRRT3
|
proline-rich transmembrane protein 3
|
|
chr2_-_86564632
|
2.949
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr3_+_186739643
|
2.932
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr8_+_17354562
|
2.898
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr5_-_16617085
|
2.842
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr2_-_86565205
|
2.806
|
NM_001164730
|
REEP1
|
receptor accessory protein 1
|
|
chr3_+_186648273
|
2.764
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr19_+_32897021
|
2.541
|
NM_207325
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr20_+_51588945
|
2.515
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr5_+_50678957
|
2.482
|
NM_002202
|
ISL1
|
ISL LIM homeobox 1
|
|
chr19_+_32896515
|
2.430
|
NM_001172774
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr12_-_94853696
|
2.428
|
NM_001042399
NM_016122
|
CCDC41
|
coiled-coil domain containing 41
|
|
chr19_-_14316980
|
2.417
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr20_+_51801821
|
2.360
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr8_+_17396285
|
2.336
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr14_-_90085325
|
2.298
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr7_+_114055048
|
2.208
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr2_+_85980600
|
2.179
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr1_-_217262946
|
2.140
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr8_-_103136486
|
2.139
|
NM_001040624
NM_001040625
NM_001040626
NM_001040627
NM_001040628
NM_001040629
|
NCALD
|
neurocalcin delta
|
|
chr2_+_97481977
|
2.126
|
NM_017623
NM_199078
|
CNNM3
|
cyclin M3
|
|
chr11_+_111807857
|
2.125
|
NM_001037954
|
DIXDC1
|
DIX domain containing 1
|
|
chr1_+_3607232
|
2.115
|
NM_001126240
NM_001126241
NM_001126242
NM_001204189
NM_001204190
NM_001204191
|
TP73
|
tumor protein p73
|
|
chr1_+_3569098
|
2.099
|
NM_001204184
NM_001204185
NM_001204186
NM_001204187
NM_001204188
NM_005427
|
TP73
|
tumor protein p73
|
|
chr8_-_103137134
|
2.070
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chrX_+_16964730
|
2.039
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr17_+_68165660
|
2.035
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr1_+_3614609
|
2.031
|
NM_001204192
|
TP73
|
tumor protein p73
|
|
chr14_-_89883306
|
2.031
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr10_-_62703822
|
2.008
|
NM_001242359
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr1_-_217311095
|
2.002
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr1_-_216896640
|
1.929
|
NM_001243515
NM_001243518
NM_001243519
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr8_-_102803438
|
1.926
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr3_+_238274
|
1.871
|
NM_001253387
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr7_-_130080823
|
1.870
|
NM_018718
|
CEP41
|
centrosomal protein 41kDa
|
|
chr15_-_49255640
|
1.824
|
NM_203349
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4
|
|
chrX_+_9431314
|
1.802
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr6_+_138483024
|
1.777
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr11_+_46299161
|
1.735
|
NM_052854
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr1_-_86043932
|
1.687
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr1_-_85930733
|
1.660
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr14_+_74111577
|
1.647
|
NM_001201366
NM_031427
|
DNAL1
|
dynein, axonemal, light chain 1
|
|
chr4_+_95679075
|
1.637
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr9_+_131217433
|
1.635
|
NM_153436
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr9_+_131219185
|
1.613
|
NM_153437
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr3_+_179370779
|
1.599
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr10_+_124768401
|
1.549
|
NM_001609
|
ACADSB
|
acyl-CoA dehydrogenase, short/branched chain
|
|
chr8_+_120885899
|
1.528
|
NM_022783
|
DEPTOR
|
DEP domain containing MTOR-interacting protein
|
|
chr19_-_33555757
|
1.488
|
NM_033103
|
RHPN2
|
rhophilin, Rho GTPase binding protein 2
|
|
chr9_-_139922705
|
1.487
|
NM_001606
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr7_+_30174343
|
1.484
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr2_-_213403280
|
1.478
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr2_-_62733475
|
1.470
|
NM_198276
|
TMEM17
|
transmembrane protein 17
|
|
chr6_+_1312674
|
1.464
|
NM_033260
|
FOXQ1
|
forkhead box Q1
|
|
chr1_-_227505796
|
1.458
|
NM_003607
NM_014826
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like)
|
|
chrX_+_9432980
|
1.455
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr9_+_131218678
|
1.444
|
NM_153433
NM_153440
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr3_-_161089747
|
1.425
|
NM_001040100
|
SPTSSB
|
serine palmitoyltransferase, small subunit B
|
|
chr5_+_153825497
|
1.419
|
NM_001131062
NM_001131063
NM_024632
|
SAP30L
|
SAP30-like
|
|
chr3_+_14444083
|
1.418
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr11_+_76777926
|
1.386
|
NM_004055
|
CAPN5
|
calpain 5
|
|
chr17_+_72322350
|
1.380
|
NM_153209
|
KIF19
|
kinesin family member 19
|
|
chr3_+_69812961
|
1.364
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr6_-_109762373
|
1.360
|
NM_001111298
NM_173672
|
PPIL6
|
peptidylprolyl isomerase (cyclophilin)-like 6
|
|
chr12_+_110906203
|
1.338
|
NM_013300
|
C12orf24
|
chromosome 12 open reading frame 24
|
|
chr21_-_34852282
|
1.337
|
NM_006134
|
TMEM50B
|
transmembrane protein 50B
|
|
chr3_-_37034609
|
1.298
|
NM_014805
|
EPM2AIP1
|
EPM2A (laforin) interacting protein 1
|
|
chrY_+_16634487
|
1.290
|
NM_001206850
|
NLGN4Y
|
neuroligin 4, Y-linked
|
|
chr16_+_14164879
|
1.266
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr9_-_139923214
|
1.229
|
NM_212533
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr1_-_234614848
|
1.213
|
NM_005646
|
TARBP1
|
TAR (HIV-1) RNA binding protein 1
|
|
chr6_+_36164528
|
1.203
|
NM_015695
|
BRPF3
|
bromodomain and PHD finger containing, 3
|
|
chr2_+_54952148
|
1.199
|
NM_001039753
|
EML6
|
echinoderm microtubule associated protein like 6
|
|
chr5_+_127419407
|
1.190
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr10_+_72163860
|
1.185
|
NM_004096
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr6_-_79787939
|
1.161
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr3_+_69915374
|
1.158
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr4_+_75858205
|
1.151
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr3_+_69788562
|
1.143
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr9_+_131218279
|
1.139
|
NM_001242352
NM_001242353
NM_001242354
NM_002540
NM_153432
NM_153435
NM_153439
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr20_-_21494663
|
1.134
|
NM_002509
|
NKX2-2
|
NK2 homeobox 2
|
|
chr3_+_23986735
|
1.126
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chrX_-_106449543
|
1.124
|
NM_017681
|
NUP62CL
|
nucleoporin 62kDa C-terminal like
|
|
chr14_-_39901619
|
1.118
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr12_-_56652118
|
1.100
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr3_+_69812620
|
1.098
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_+_23987514
|
1.096
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr2_-_152955502
|
1.095
|
NM_000726
NM_001145798
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr2_+_61292999
|
1.083
|
NM_001129993
|
KIAA1841
|
KIAA1841
|
|
chr7_+_17338182
|
1.080
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr16_-_19896105
|
1.069
|
NM_016235
|
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B
|
|
chr1_+_109656570
|
1.066
|
NM_020775
|
KIAA1324
|
KIAA1324
|
|
chr8_-_93978264
|
1.055
|
NM_001191035
NM_001171795
NM_001171796
NM_001171797
NM_001171798
NM_001171799
|
C8orf83
|
chromosome 8 open reading frame 83
|
|
chrX_+_77166178
|
1.037
|
NM_000052
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr1_+_57110745
|
1.029
|
NM_006252
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chr14_-_65438794
|
1.028
|
NM_198686
|
RAB15
|
RAB15, member RAS onocogene family
|
|
chr14_+_64682671
|
1.023
|
NM_182910
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr2_+_45878912
|
1.018
|
NM_005400
|
PRKCE
|
protein kinase C, epsilon
|
|
chrX_+_17393537
|
1.016
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr2_-_152955208
|
1.015
|
NM_001005746
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr22_-_44893903
|
1.013
|
NM_032287
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr9_+_115983807
|
1.004
|
NM_001859
|
SLC31A1
|
solute carrier family 31 (copper transporters), member 1
|
|
chrX_+_17653412
|
0.991
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr14_+_105331581
|
0.981
|
NM_001112726
NM_015005
|
KIAA0284
|
KIAA0284
|
|
chr3_-_133614421
|
0.972
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr2_-_44587812
|
0.969
|
NM_006036
|
PREPL
|
prolyl endopeptidase-like
|
|
chr3_-_46037299
|
0.962
|
NM_024513
|
FYCO1
|
FYVE and coiled-coil domain containing 1
|
|
chr5_-_179498995
|
0.956
|
NM_018434
|
RNF130
|
ring finger protein 130
|
|
chr6_-_80247103
|
0.952
|
NM_001122769
NM_181714
|
LCA5
|
Leber congenital amaurosis 5
|
|
chr6_+_80340971
|
0.950
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr10_+_104613966
|
0.935
|
NM_001136200
NM_144591
|
C10orf32-AS3MT
C10orf32
|
C10orf32-AS3MT readthrough
chromosome 10 open reading frame 32
|
|
chr5_-_9546157
|
0.929
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chrY_+_16635625
|
0.921
|
NM_014893
|
NLGN4Y
|
neuroligin 4, Y-linked
|
|
chr9_+_976963
|
0.917
|
NM_021240
|
DMRT3
|
doublesex and mab-3 related transcription factor 3
|
|
chr17_-_1532109
|
0.914
|
NM_152346
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr5_+_131705353
|
0.905
|
NM_003060
|
SLC22A5
|
solute carrier family 22 (organic cation/carnitine transporter), member 5
|
|
chr3_+_181429711
|
0.905
|
NM_003106
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr10_-_62149487
|
0.895
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr10_-_61900659
|
0.894
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr7_-_25164902
|
0.889
|
NM_018947
|
CYCS
|
cytochrome c, somatic
|
|
chr2_-_44586854
|
0.879
|
NM_001042385
NM_001042386
|
PREPL
|
prolyl endopeptidase-like
|
|
chr7_-_112726143
|
0.877
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chrX_-_34675366
|
0.864
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr5_-_90679115
|
0.862
|
NM_020801
|
ARRDC3
|
arrestin domain containing 3
|
|
chr19_-_33793429
|
0.862
|
NM_004364
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha
|
|
chrX_+_72782961
|
0.861
|
NM_001039840
|
CHIC1
|
cysteine-rich hydrophobic domain 1
|
|
chr10_-_116418056
|
0.860
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr14_-_81405883
|
0.853
|
NM_152446
|
CEP128
|
centrosomal protein 128kDa
|
|
chr20_+_278200
|
0.849
|
NM_033089
|
ZCCHC3
|
zinc finger, CCHC domain containing 3
|
|
chr10_-_116444374
|
0.844
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr10_-_62332666
|
0.836
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr14_+_64680858
|
0.834
|
NM_182913
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr9_+_119449573
|
0.830
|
NM_001099679
NM_012210
|
TRIM32
|
tripartite motif containing 32
|
|
chr7_-_47621741
|
0.826
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr2_-_158731622
|
0.822
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr18_-_47721165
|
0.818
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr22_-_39548448
|
0.817
|
NM_175709
|
CBX7
|
chromobox homolog 7
|
|
chr10_-_62493282
|
0.814
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr8_+_28351694
|
0.813
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr8_+_101170510
|
0.810
|
NM_003114
|
SPAG1
|
sperm associated antigen 1
|
|
chr13_+_115079943
|
0.809
|
NM_001164144
NM_001164145
NM_032436
|
CHAMP1
|
chromosome alignment maintaining phosphoprotein 1
|
|
chr8_+_110346549
|
0.798
|
NM_001193557
NM_020189
|
ENY2
|
enhancer of yellow 2 homolog (Drosophila)
|
|
chr12_+_52345437
|
0.797
|
NM_004302
NM_020328
|
ACVR1B
|
activin A receptor, type IB
|
|
chr14_+_58765209
|
0.794
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr9_-_80646150
|
0.794
|
NM_002072
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide
|
|
chr8_+_101170231
|
0.790
|
NM_172218
|
SPAG1
|
sperm associated antigen 1
|
|
chr10_-_116286661
|
0.780
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr2_-_152830448
|
0.778
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr11_-_79151694
|
0.773
|
NM_001098816
|
ODZ4
|
odz, odd Oz/ten-m homolog 4 (Drosophila)
|
|
chrX_+_70752911
|
0.768
|
NM_181672
NM_181673
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr7_-_95225802
|
0.768
|
NM_002612
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4
|
|
chr14_+_56046796
|
0.766
|
NM_001079521
NM_001079522
NM_004986
NM_182926
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chr12_-_106533810
|
0.763
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr10_-_61469322
|
0.760
|
NM_194298
|
LOC100129721
SLC16A9
|
uncharacterized LOC100129721
solute carrier family 16, member 9 (monocarboxylic acid transporter 9)
|
|
chr17_-_27621163
|
0.757
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr6_+_52285804
|
0.754
|
NM_001172420
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chr9_+_79056581
|
0.751
|
NM_001097634
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr4_-_88141614
|
0.745
|
NM_020803
|
KLHL8
|
kelch-like 8 (Drosophila)
|
|
chr7_+_94536926
|
0.740
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr9_+_71320105
|
0.736
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr9_-_79520878
|
0.735
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr4_+_108745718
|
0.732
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr10_+_92980322
|
0.726
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr20_-_50384899
|
0.725
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr4_-_16228097
|
0.718
|
NM_153365
|
TAPT1
|
transmembrane anterior posterior transformation 1
|
|
chr7_+_43966007
|
0.713
|
NM_015983
|
UBE2D4
|
ubiquitin-conjugating enzyme E2D 4 (putative)
|
|
chr2_+_39103102
|
0.710
|
NM_001145450
|
MORN2
|
MORN repeat containing 2
|
|
chr7_-_112727832
|
0.704
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr3_+_69985750
|
0.703
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr1_-_202858225
|
0.700
|
NM_002871
|
RABIF
|
RAB interacting factor
|
|
chr9_+_4490193
|
0.699
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr4_+_108814419
|
0.698
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr2_-_218808705
|
0.695
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr6_+_30295007
|
0.688
|
NM_172016
|
TRIM39
|
tripartite motif containing 39
|
|
chr2_-_158732341
|
0.682
|
NM_001111067
|
ACVR1
|
activin A receptor, type I
|
|
chr4_-_37687932
|
0.679
|
NM_001085399
NM_001085400
|
RELL1
|
RELT-like 1
|
|
chr2_-_174828775
|
0.675
|
NM_001017371
|
SP3
|
Sp3 transcription factor
|
|
chr6_+_18155535
|
0.664
|
NM_153042
|
KDM1B
|
lysine (K)-specific demethylase 1B
|
|
chr8_+_92082423
|
0.662
|
NM_016023
|
OTUD6B
|
OTU domain containing 6B
|
|
chr17_+_61699786
|
0.661
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr2_-_68479481
|
0.657
|
NM_000945
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha
|
|
chr2_-_96931744
|
0.651
|
NM_001193304
NM_017849
|
TMEM127
|
transmembrane protein 127
|
|
chr6_-_90121966
|
0.644
|
NM_021244
|
RRAGD
|
Ras-related GTP binding D
|
|
chr5_+_67511578
|
0.637
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr2_+_32288579
|
0.637
|
NM_014946
NM_199436
|
SPAST
|
spastin
|
|
chr12_-_56727446
|
0.637
|
NM_001127460
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|