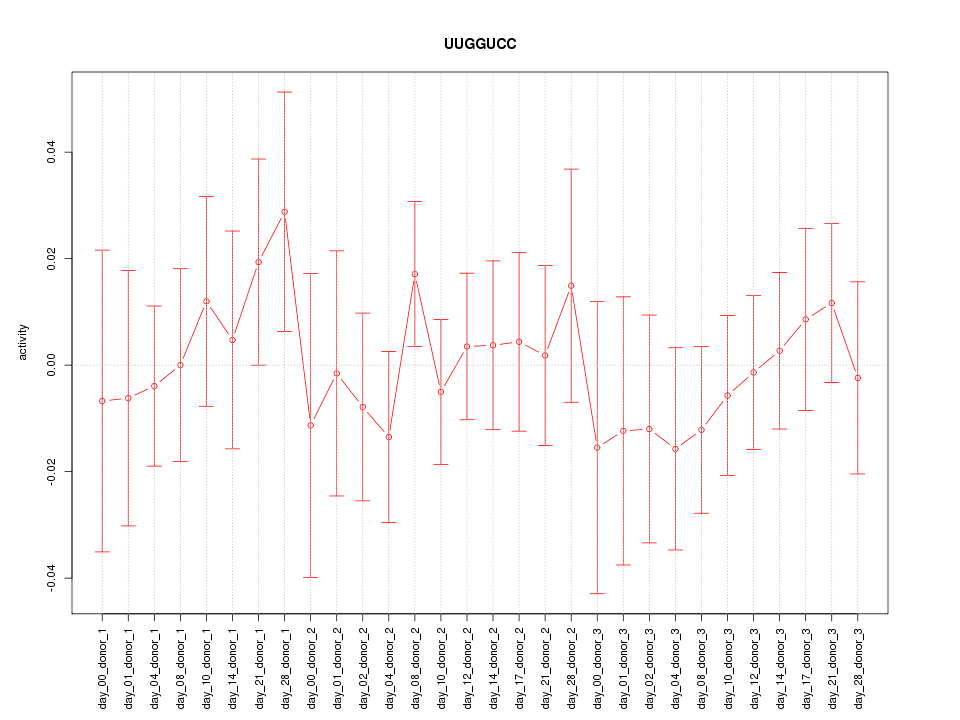
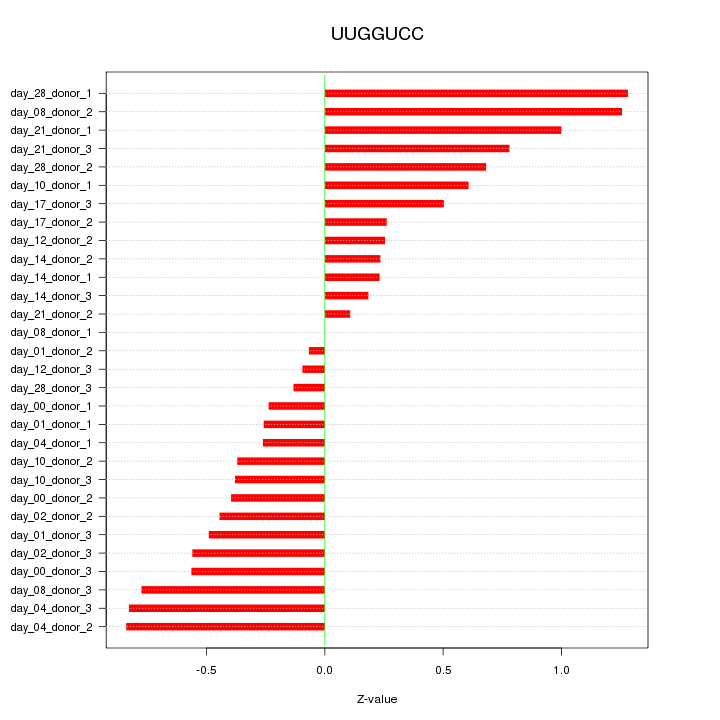
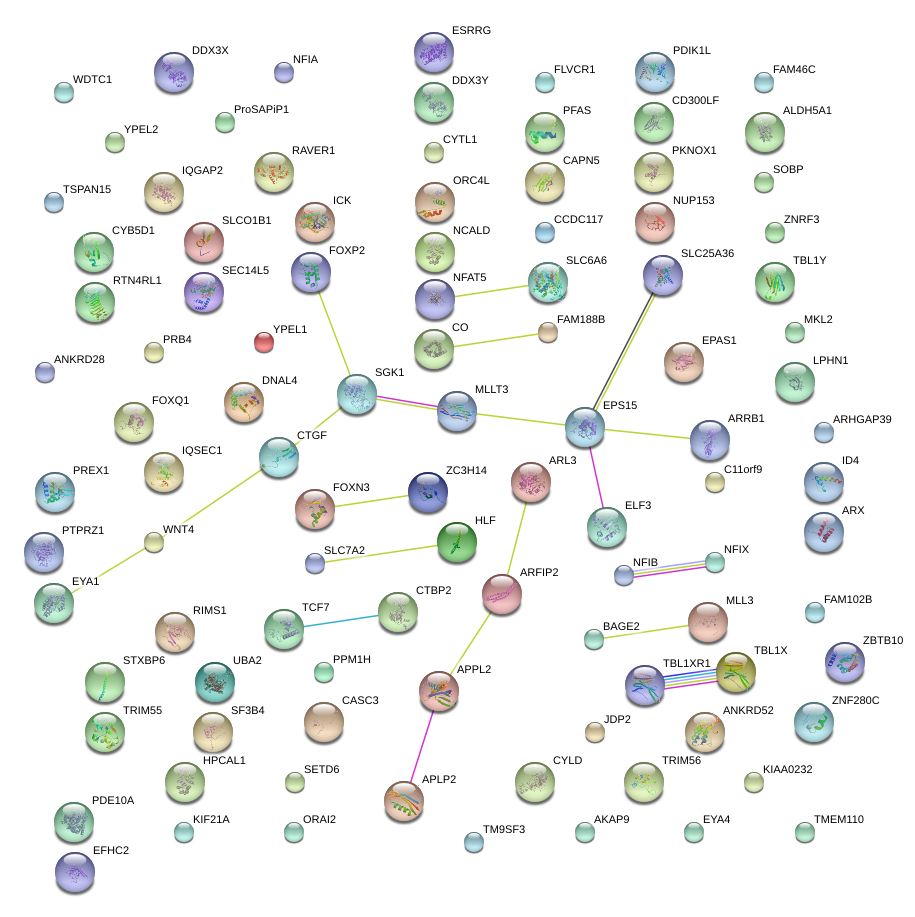
|
chr6_-_132272311
|
3.124
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr8_+_17354562
|
1.813
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr1_+_118148506
|
1.812
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr8_+_17396285
|
1.490
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr1_-_22469414
|
1.408
|
NM_030761
|
WNT4
|
wingless-type MMTV integration site family, member 4
|
|
chr7_+_121513158
|
1.288
|
NM_001206838
NM_001206839
NM_002851
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1
|
|
chr6_+_19837599
|
1.240
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr16_+_58549382
|
1.059
|
NM_001160305
NM_024860
|
SETD6
|
SET domain containing 6
|
|
chr7_+_114055048
|
1.057
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr12_-_63328663
|
0.993
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr12_-_39836760
|
0.914
|
NM_001173463
NM_001173464
NM_001173465
NM_017641
|
KIF21A
|
kinesin family member 21A
|
|
chr1_+_61547625
|
0.897
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr8_-_145838887
|
0.877
|
NM_025251
|
ARHGAP39
|
Rho GTPase activating protein 39
|
|
chr8_+_81398416
|
0.871
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr14_-_90085325
|
0.863
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr1_+_61542928
|
0.854
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr22_+_29279573
|
0.824
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr7_+_100728719
|
0.788
|
NM_030961
|
TRIM56
|
tripartite motif containing 56
|
|
chr6_+_107811272
|
0.766
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr11_+_76777926
|
0.766
|
NM_004055
|
CAPN5
|
calpain 5
|
|
chr20_-_3149070
|
0.760
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr17_+_53342320
|
0.745
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr14_-_89883306
|
0.737
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chrX_+_9431314
|
0.731
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr7_+_30960914
|
0.716
|
NM_001185060
NM_001185061
NM_001185062
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr6_+_24495166
|
0.702
|
NM_001080
NM_170740
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1
|
|
chr1_+_61547388
|
0.670
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr7_+_102073953
|
0.666
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr9_-_20622476
|
0.655
|
NM_004529
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr17_-_1928177
|
0.632
|
NM_178568
|
RTN4RL1
|
reticulon 4 receptor-like 1
|
|
chr3_-_15900928
|
0.630
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr1_-_217262946
|
0.617
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chrY_+_15016018
|
0.595
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr6_+_133562488
|
0.592
|
NM_004100
NM_172103
NM_172105
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr1_-_217311095
|
0.588
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr6_+_72926462
|
0.581
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_+_213031596
|
0.575
|
NM_014053
|
FLVCR1
|
feline leukemia virus subgroup C cellular receptor 1
|
|
chrX_+_9432980
|
0.570
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr1_-_216896640
|
0.562
|
NM_001243515
NM_001243518
NM_001243519
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr15_-_77712436
|
0.561
|
NM_024776
|
PEAK1
|
NKF3 kinase family member
|
|
chr4_+_6784453
|
0.550
|
NM_001100590
NM_014743
|
KIAA0232
|
KIAA0232
|
|
chr16_+_14164879
|
0.546
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr7_+_91570150
|
0.540
|
NM_005751
NM_147185
|
AKAP9
|
A kinase (PRKA) anchor protein (yotiao) 9
|
|
chr6_-_134497069
|
0.533
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr17_+_38296506
|
0.528
|
NM_007359
|
CASC3
|
cancer susceptibility candidate 3
|
|
chr6_-_134496006
|
0.522
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr7_+_30951414
|
0.518
|
NM_198098
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr8_-_72268720
|
0.516
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr11_-_75062650
|
0.511
|
NM_004041
NM_020251
|
ARRB1
|
arrestin, beta 1
|
|
chrX_-_129402753
|
0.511
|
NM_017666
|
ZNF280C
|
zinc finger protein 280C
|
|
chr3_+_14444083
|
0.503
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr10_-_98346793
|
0.502
|
NM_020123
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr6_+_72922513
|
0.502
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr14_-_25519076
|
0.496
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr6_+_1312674
|
0.492
|
NM_033260
|
FOXQ1
|
forkhead box Q1
|
|
chr8_+_67039130
|
0.488
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr19_-_14316980
|
0.486
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr10_+_71211215
|
0.486
|
NM_012339
|
TSPAN15
|
tetraspanin 15
|
|
chr22_+_29313376
|
0.472
|
NM_032173
|
ZNRF3
|
zinc and ring finger 3
|
|
chr16_+_50775960
|
0.463
|
NM_001042355
NM_015247
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr16_+_69599868
|
0.463
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr16_+_5008317
|
0.461
|
NM_014692
|
SEC14L5
|
SEC14-like 5 (S. cerevisiae)
|
|
chr6_+_72926144
|
0.449
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr7_-_152133071
|
0.439
|
NM_170606
|
MLL3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr3_-_15839674
|
0.438
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr22_+_29168628
|
0.434
|
NM_173510
|
CCDC117
|
coiled-coil domain containing 117
|
|
chrX_-_44202834
|
0.433
|
NM_025184
|
EFHC2
|
EF-hand domain (C-terminal) containing 2
|
|
chr11_+_61520000
|
0.432
|
NM_001127392
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr8_-_103136486
|
0.429
|
NM_001040624
NM_001040625
NM_001040626
NM_001040627
NM_001040628
NM_001040629
|
NCALD
|
neurocalcin delta
|
|
chr17_+_57408907
|
0.428
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr1_+_109102970
|
0.426
|
NM_001010883
|
FAM102B
|
family with sequence similarity 102, member B
|
|
chr5_+_75699003
|
0.426
|
NM_006633
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr8_-_72274466
|
0.426
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr12_-_56652118
|
0.424
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr19_-_10444186
|
0.416
|
NM_133452
|
RAVER1
|
ribonucleoprotein, PTB-binding 1
|
|
chr17_+_7761063
|
0.415
|
NM_144607
|
CYB5D1
|
cytochrome b5 domain containing 1
|
|
chrY_+_15016698
|
0.414
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr20_-_47444404
|
0.413
|
NM_020820
|
PREX1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1
|
|
chr17_+_8152594
|
0.413
|
NM_012393
|
PFAS
|
phosphoribosylformylglycinamidine synthase
|
|
chr8_-_103137134
|
0.411
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chr11_-_6502563
|
0.409
|
NM_001242854
NM_001242855
NM_001242856
NM_012402
|
ARFIP2
|
ADP-ribosylation factor interacting protein 2
|
|
chr16_+_50776012
|
0.409
|
NM_001042412
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr3_-_13114547
|
0.405
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr6_-_134639195
|
0.397
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr12_-_105629790
|
0.391
|
NM_001251904
NM_018171
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2
|
|
chr1_+_201979689
|
0.390
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr6_-_134498973
|
0.389
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr8_-_102803438
|
0.388
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr5_+_133451297
|
0.386
|
NM_001134851
NM_201632
NM_201634
NM_213648
|
TCF7
|
transcription factor 7 (T-cell specific, HMG-box)
|
|
chr6_-_52926588
|
0.381
|
NM_014920
NM_016513
|
ICK
|
intestinal cell (MAK-like) kinase
|
|
chr21_+_44394634
|
0.381
|
NM_004571
|
PKNOX1
|
PBX/knotted 1 homeobox 1
|
|
chr1_+_27561006
|
0.372
|
NM_015023
|
WDTC1
|
WD and tetratricopeptide repeats 1
|
|
chr3_+_140660639
|
0.369
|
NM_001104647
NM_018155
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr14_+_75894919
|
0.369
|
NM_001135048
NM_130469
|
JDP2
|
Jun dimerization protein 2
|
|
chr6_+_72596648
|
0.369
|
NM_014989
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_-_17706617
|
0.363
|
NM_005124
|
NUP153
|
nucleoporin 153kDa
|
|
chr14_+_89060729
|
0.363
|
NM_207662
|
ZC3H14
|
zinc finger CCCH-type containing 14
|
|
chr6_-_166075520
|
0.362
|
NM_006661
NM_001130690
|
PDE10A
|
phosphodiesterase 10A
|
|
chr17_-_72709010
|
0.361
|
NM_139018
|
CD300LF
|
CD300 molecule-like family member f
|
|
chr1_-_51984994
|
0.358
|
NM_001981
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr3_-_52931595
|
0.352
|
NM_001198974
NM_198563
|
TMEM110-MUSTN1
TMEM110
|
TMEM110-MUSTN1 readthrough
transmembrane protein 110
|
|
chrX_-_25034064
|
0.351
|
NM_139058
|
ARX
|
aristaless related homeobox
|
|
chr1_-_149900143
|
0.342
|
NM_005850
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa
|
|
chr10_-_104473957
|
0.341
|
NM_004311
|
ARL3
|
ADP-ribosylation factor-like 3
|
|
chr5_+_133450353
|
0.339
|
NM_003202
|
TCF7
|
transcription factor 7 (T-cell specific, HMG-box)
|
|
chr14_+_89029252
|
0.339
|
NM_001160103
NM_001160104
NM_024824
NM_207660
|
ZC3H14
|
zinc finger CCCH-type containing 14
|
|
chr2_-_148779172
|
0.335
|
NM_181742
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr1_+_26438267
|
0.335
|
NM_001243533
NM_152835
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr22_-_39190146
|
0.332
|
NM_005740
|
DNAL4
|
dynein, axonemal, light chain 4
|
|
chr10_-_126849556
|
0.331
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr9_-_139922705
|
0.329
|
NM_001606
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr14_+_103243813
|
0.325
|
NM_001199427
NM_003300
NM_145725
NM_145726
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr4_-_140060601
|
0.324
|
NM_201999
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr4_-_140005464
|
0.324
|
NM_006874
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr9_+_2015218
|
0.322
|
NM_003070
NM_139045
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr14_+_89029690
|
0.321
|
NM_207661
|
ZC3H14
|
zinc finger CCCH-type containing 14
|
|
chr8_-_93978264
|
0.321
|
NM_001191035
NM_001171795
NM_001171796
NM_001171797
NM_001171798
NM_001171799
|
C8orf83
|
chromosome 8 open reading frame 83
|
|
chr17_+_61699786
|
0.314
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr7_-_138665853
|
0.311
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr2_-_174828775
|
0.310
|
NM_001017371
|
SP3
|
Sp3 transcription factor
|
|
chr6_+_73076431
|
0.310
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr2_-_16847070
|
0.304
|
NM_030797
|
FAM49A
|
family with sequence similarity 49, member A
|
|
chr10_+_60272868
|
0.302
|
NM_001080512
|
BICC1
|
bicaudal C homolog 1 (Drosophila)
|
|
chr11_+_61522860
|
0.301
|
NM_013279
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr19_-_7293876
|
0.301
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr10_+_69644938
|
0.301
|
NM_001142498
|
SIRT1
|
sirtuin 1
|
|
chr9_-_139923214
|
0.297
|
NM_212533
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr4_+_108745718
|
0.295
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr3_-_107809754
|
0.295
|
NM_001777
NM_198793
|
CD47
|
CD47 molecule
|
|
chr16_-_68482373
|
0.295
|
NM_018667
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II)
|
|
chr15_-_37392358
|
0.294
|
NM_170674
NM_170675
NM_170676
NM_170677
|
MEIS2
|
Meis homeobox 2
|
|
chr4_+_26321307
|
0.293
|
NM_005349
NM_203283
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr1_+_11333244
|
0.290
|
NM_013319
|
UBIAD1
|
UbiA prenyltransferase domain containing 1
|
|
chr6_-_109330190
|
0.290
|
NM_001199934
|
SESN1
|
sestrin 1
|
|
chr18_+_9137560
|
0.289
|
NM_001204056
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr22_-_17602142
|
0.288
|
NM_031890
NM_001163079
|
CECR6
|
cat eye syndrome chromosome region, candidate 6
|
|
chr16_+_50186809
|
0.284
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr17_-_6459746
|
0.281
|
NM_001165966
NM_031220
|
PITPNM3
|
PITPNM family member 3
|
|
chr15_-_43877043
|
0.278
|
NM_014659
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1
|
|
chr2_-_148778262
|
0.277
|
NM_001190882
NM_001190879
NM_002552
NM_001190881
NM_181741
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr4_-_39640461
|
0.274
|
NM_174921
|
C4orf34
|
chromosome 4 open reading frame 34
|
|
chr15_-_37393364
|
0.273
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr2_-_27886443
|
0.273
|
NM_014860
|
SUPT7L
|
suppressor of Ty 7 (S. cerevisiae)-like
|
|
chr6_-_109415514
|
0.271
|
NM_014454
|
SESN1
|
sestrin 1
|
|
chr15_-_37391596
|
0.269
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr1_+_167189948
|
0.268
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr18_+_905296
|
0.267
|
NM_001117
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1 (pituitary)
|
|
chr12_-_42538432
|
0.266
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr7_-_73184535
|
0.265
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr13_+_98795410
|
0.265
|
NM_001001715
NM_005766
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr15_-_43882387
|
0.264
|
NM_001130858
NM_001130859
NM_001190214
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1
|
|
chr20_-_41818464
|
0.264
|
NM_007050
NM_133170
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chr17_+_7608413
|
0.262
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr22_+_40440664
|
0.262
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr15_-_37390372
|
0.262
|
NM_172315
|
MEIS2
|
Meis homeobox 2
|
|
chr14_-_23388380
|
0.261
|
NM_001077351
NM_001077352
NM_018107
|
RBM23
|
RNA binding motif protein 23
|
|
chr6_-_26659913
|
0.260
|
NM_001242797
NM_001242798
NM_001242799
NM_024639
|
ZNF322
|
zinc finger protein 322
|
|
chr5_-_79286793
|
0.257
|
NM_001010891
NM_001167741
|
MTX3
|
metaxin 3
|
|
chr9_+_214860
|
0.257
|
NM_203447
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr4_+_37246689
|
0.257
|
NM_001144990
|
KIAA1239
|
KIAA1239
|
|
chr6_-_143266283
|
0.256
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr4_+_108814419
|
0.255
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr3_-_133614421
|
0.250
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr17_-_10633645
|
0.246
|
NM_001004313
|
TMEM220
|
transmembrane protein 220
|
|
chr3_+_150804675
|
0.245
|
NM_053002
|
MED12L
|
mediator complex subunit 12-like
|
|
chr14_+_75894508
|
0.243
|
NM_001135047
|
JDP2
|
Jun dimerization protein 2
|
|
chr12_-_77459198
|
0.243
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr12_+_124457568
|
0.242
|
NM_001204299
NM_001204298
NM_152437
|
ZNF664-FAM101A
ZNF664
|
protein FAM101A
zinc finger protein 664
|
|
chr1_-_212004113
|
0.240
|
NM_014873
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chr10_+_69644341
|
0.239
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr5_+_175665337
|
0.238
|
NM_198567
|
C5orf25
|
chromosome 5 open reading frame 25
|
|
chr7_+_43152196
|
0.238
|
NM_015052
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1
|
|
chr14_-_78083109
|
0.237
|
NM_004863
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2
|
|
chr1_-_205782135
|
0.236
|
NM_173854
|
SLC41A1
|
solute carrier family 41, member 1
|
|
chr22_-_41842853
|
0.234
|
NM_016272
|
TOB2
|
transducer of ERBB2, 2
|
|
chrX_-_77041678
|
0.234
|
NM_000489
NM_138270
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr9_+_273025
|
0.232
|
NM_001190458
NM_001193536
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr7_-_37488554
|
0.232
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr14_+_90863302
|
0.232
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr7_-_100286869
|
0.231
|
NM_022574
|
GIGYF1
|
GRB10 interacting GYF protein 1
|
|
chr6_-_79787939
|
0.229
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr6_-_109330752
|
0.228
|
NM_001199933
|
SESN1
|
sestrin 1
|
|
chr5_-_37839781
|
0.228
|
NM_000514
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr17_+_73780842
|
0.228
|
NM_001080419
|
UNK
|
unkempt homolog (Drosophila)
|
|
chr11_+_120110902
|
0.227
|
NM_014352
|
POU2F3
|
POU class 2 homeobox 3
|
|
chr10_-_1779669
|
0.225
|
NM_018702
|
ADARB2
|
adenosine deaminase, RNA-specific, B2
|
|
chr3_+_43328001
|
0.224
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr5_+_43121641
|
0.223
|
NM_003432
|
ZNF131
|
zinc finger protein 131
|
|
chr17_+_29718641
|
0.222
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr14_+_75898836
|
0.221
|
NM_001135049
|
JDP2
|
Jun dimerization protein 2
|
|
chr11_+_22688156
|
0.221
|
NM_001143830
|
GAS2
|
growth arrest-specific 2
|
|
chr10_-_21463019
|
0.221
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr7_-_37024664
|
0.220
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr9_+_133971862
|
0.220
|
NM_001185095
NM_001185096
NM_031426
|
AIF1L
|
allograft inflammatory factor 1-like
|
|
chr22_-_39268230
|
0.217
|
NM_014292
|
CBX6
|
chromobox homolog 6
|
|
chr3_-_15798057
|
0.216
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr8_-_38325241
|
0.214
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr3_-_148804122
|
0.214
|
NM_003071
NM_139048
|
HLTF
|
helicase-like transcription factor
|
|
chr1_+_167298280
|
0.212
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr17_+_7788094
|
0.212
|
NM_001005271
|
CHD3
|
chromodomain helicase DNA binding protein 3
|