Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
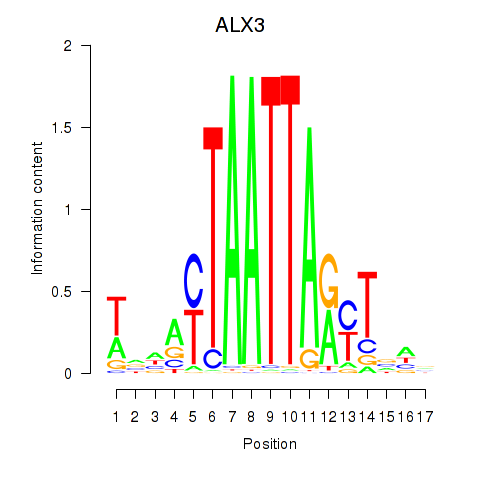
Results for ALX3
Z-value: 0.66
Transcription factors associated with ALX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ALX3
|
ENSG00000156150.6 | ALX homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ALX3 | hg19_v2_chr1_-_110613276_110613322 | -0.19 | 3.0e-01 | Click! |
Activity profile of ALX3 motif
Sorted Z-values of ALX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_25348007 | 4.24 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr5_-_35938674 | 3.52 |
ENST00000397366.1
ENST00000513623.1 ENST00000514524.1 ENST00000397367.2 |
CAPSL
|
calcyphosine-like |
| chr11_+_101918153 | 3.43 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr12_+_7013897 | 3.16 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr12_+_7014064 | 3.12 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr12_+_7014126 | 2.32 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr12_-_10282836 | 2.14 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr11_-_63376013 | 2.03 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr16_-_55866997 | 1.75 |
ENST00000360526.3
ENST00000361503.4 |
CES1
|
carboxylesterase 1 |
| chr11_+_71900703 | 1.73 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr9_-_117111222 | 1.72 |
ENST00000374079.4
|
AKNA
|
AT-hook transcription factor |
| chr11_+_71900572 | 1.70 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr12_-_10282681 | 1.59 |
ENST00000533022.1
|
CLEC7A
|
C-type lectin domain family 7, member A |
| chr16_-_28634874 | 1.57 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr12_-_10282742 | 1.55 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr4_+_86525299 | 1.44 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr13_-_86373536 | 1.40 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr14_-_54423529 | 1.28 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr1_+_47533160 | 1.27 |
ENST00000334194.3
|
CYP4Z1
|
cytochrome P450, family 4, subfamily Z, polypeptide 1 |
| chr4_-_70518941 | 1.27 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr16_-_28608364 | 1.26 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr16_-_28608424 | 1.15 |
ENST00000335715.4
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr16_-_28621353 | 1.14 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr4_-_25865159 | 1.12 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr17_+_35851570 | 1.11 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr1_-_150738261 | 1.04 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr19_+_50016411 | 0.99 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr3_+_140396881 | 0.97 |
ENST00000286349.3
|
TRIM42
|
tripartite motif containing 42 |
| chr16_-_28937027 | 0.95 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr16_-_28621312 | 0.95 |
ENST00000314752.7
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr19_+_50016610 | 0.94 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr19_+_49199209 | 0.93 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr11_+_100862811 | 0.88 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr17_+_1674982 | 0.82 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr8_+_105235572 | 0.82 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr14_+_74034310 | 0.82 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr9_+_90112767 | 0.81 |
ENST00000408954.3
|
DAPK1
|
death-associated protein kinase 1 |
| chr7_+_48211048 | 0.79 |
ENST00000435803.1
|
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr1_+_28261492 | 0.77 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr10_+_695888 | 0.74 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr16_-_28621298 | 0.73 |
ENST00000566189.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr4_-_138453559 | 0.72 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr1_+_28261621 | 0.71 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr9_+_90112741 | 0.69 |
ENST00000469640.2
|
DAPK1
|
death-associated protein kinase 1 |
| chrX_+_77154935 | 0.69 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr9_+_90112590 | 0.68 |
ENST00000472284.1
|
DAPK1
|
death-associated protein kinase 1 |
| chr9_+_90112117 | 0.64 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr12_+_131438443 | 0.62 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr11_+_6897856 | 0.61 |
ENST00000379829.2
|
OR10A4
|
olfactory receptor, family 10, subfamily A, member 4 |
| chr10_-_28571015 | 0.61 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr6_+_26402465 | 0.60 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr5_-_20575959 | 0.59 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr13_+_24144796 | 0.56 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr2_+_217524323 | 0.55 |
ENST00000456764.1
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr4_+_37455536 | 0.55 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr5_-_96478466 | 0.54 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr3_-_191000172 | 0.54 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr3_+_195447738 | 0.53 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr19_+_42580274 | 0.52 |
ENST00000359044.4
|
ZNF574
|
zinc finger protein 574 |
| chr7_-_111032971 | 0.52 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr16_+_31885079 | 0.52 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr11_+_57480046 | 0.51 |
ENST00000378312.4
ENST00000278422.4 |
TMX2
|
thioredoxin-related transmembrane protein 2 |
| chr15_-_98417780 | 0.50 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr1_+_12976450 | 0.49 |
ENST00000361079.2
|
PRAMEF7
|
PRAME family member 7 |
| chr13_+_78315295 | 0.48 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr6_+_26402517 | 0.47 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr18_-_53303123 | 0.47 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr1_+_117544366 | 0.45 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr14_-_78083112 | 0.45 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr1_-_190446759 | 0.45 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr5_+_140557371 | 0.44 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr12_-_46121554 | 0.42 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr16_+_31724552 | 0.42 |
ENST00000539915.1
ENST00000316491.9 ENST00000399681.3 ENST00000398696.3 ENST00000534369.1 |
ZNF720
|
zinc finger protein 720 |
| chr22_-_32651326 | 0.42 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr8_-_102803163 | 0.42 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chrX_+_130192318 | 0.41 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr6_+_26440700 | 0.40 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr11_+_7618413 | 0.40 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr4_-_66536196 | 0.39 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr1_-_54411240 | 0.39 |
ENST00000371378.2
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr1_-_150669500 | 0.39 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr1_-_54411255 | 0.39 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr2_-_43266680 | 0.39 |
ENST00000425212.1
ENST00000422351.1 ENST00000449766.1 |
AC016735.2
|
AC016735.2 |
| chr4_+_86699834 | 0.38 |
ENST00000395183.2
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr5_-_94417339 | 0.38 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr6_+_52442083 | 0.38 |
ENST00000606714.1
|
TRAM2-AS1
|
TRAM2 antisense RNA 1 (head to head) |
| chr7_+_133812052 | 0.37 |
ENST00000285928.2
|
LRGUK
|
leucine-rich repeats and guanylate kinase domain containing |
| chr20_-_29978286 | 0.37 |
ENST00000376315.2
|
DEFB119
|
defensin, beta 119 |
| chr4_+_113568207 | 0.37 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr1_+_160370344 | 0.37 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr11_-_128894053 | 0.36 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chrX_-_13835147 | 0.35 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr15_-_89755034 | 0.34 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr9_+_105757590 | 0.34 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr1_+_225600404 | 0.33 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr20_-_50418947 | 0.32 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr3_+_138340049 | 0.32 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr20_-_50418972 | 0.31 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr2_+_103089756 | 0.30 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chrX_+_36254051 | 0.30 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr14_+_39736582 | 0.29 |
ENST00000556148.1
ENST00000348007.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr19_+_47421933 | 0.29 |
ENST00000404338.3
|
ARHGAP35
|
Rho GTPase activating protein 35 |
| chr4_-_120243545 | 0.28 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr17_-_39341594 | 0.27 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr16_-_66584059 | 0.26 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr17_-_4938712 | 0.26 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr7_-_43965937 | 0.26 |
ENST00000455877.1
ENST00000223341.7 ENST00000447717.3 ENST00000426198.1 |
URGCP
|
upregulator of cell proliferation |
| chr2_+_234826016 | 0.26 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr3_+_138340067 | 0.25 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr15_+_44719970 | 0.25 |
ENST00000558966.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr12_+_8666126 | 0.25 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr2_-_152118352 | 0.25 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr16_+_53133070 | 0.24 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr7_+_138145076 | 0.24 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr11_+_92702886 | 0.24 |
ENST00000257068.2
ENST00000528076.1 |
MTNR1B
|
melatonin receptor 1B |
| chr12_+_66582919 | 0.24 |
ENST00000545837.1
ENST00000457197.2 |
IRAK3
|
interleukin-1 receptor-associated kinase 3 |
| chrX_+_108779004 | 0.24 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr4_-_66536057 | 0.23 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr10_+_91152303 | 0.23 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr5_+_176811431 | 0.23 |
ENST00000512593.1
ENST00000324417.5 |
SLC34A1
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr17_-_39191107 | 0.22 |
ENST00000344363.5
|
KRTAP1-3
|
keratin associated protein 1-3 |
| chr14_+_55493920 | 0.22 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr8_+_107738240 | 0.22 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr9_-_5339873 | 0.22 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr2_+_135596180 | 0.21 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr5_+_140201183 | 0.21 |
ENST00000529619.1
ENST00000529859.1 ENST00000378126.3 |
PCDHA5
|
protocadherin alpha 5 |
| chr3_-_160823040 | 0.21 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr5_+_140762268 | 0.21 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr8_-_93978357 | 0.21 |
ENST00000522925.1
ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr3_-_160823158 | 0.20 |
ENST00000392779.2
ENST00000392780.1 ENST00000494173.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr2_+_113299990 | 0.20 |
ENST00000537335.1
ENST00000417433.2 |
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr15_-_75748115 | 0.20 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr12_-_122985067 | 0.20 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr14_+_55494323 | 0.20 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr2_-_89340242 | 0.20 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr7_-_149158187 | 0.20 |
ENST00000247930.4
|
ZNF777
|
zinc finger protein 777 |
| chr17_+_18086392 | 0.19 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr6_-_32095968 | 0.19 |
ENST00000375203.3
ENST00000375201.4 |
ATF6B
|
activating transcription factor 6 beta |
| chr5_+_140227357 | 0.19 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr2_+_135596106 | 0.19 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr9_+_12775011 | 0.19 |
ENST00000319264.3
|
LURAP1L
|
leucine rich adaptor protein 1-like |
| chr15_-_37393406 | 0.18 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr11_+_5710919 | 0.18 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr2_-_231860596 | 0.18 |
ENST00000441063.1
ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2
|
SPATA3 antisense RNA 1 (head to head) |
| chr14_+_22977587 | 0.17 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr7_+_43966018 | 0.16 |
ENST00000222402.3
ENST00000446008.1 ENST00000394798.4 |
UBE2D4
|
ubiquitin-conjugating enzyme E2D 4 (putative) |
| chr1_-_150669604 | 0.16 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr3_+_186353756 | 0.16 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr10_+_90660832 | 0.16 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr11_-_13011081 | 0.16 |
ENST00000532541.1
ENST00000526388.1 ENST00000534477.1 ENST00000531402.1 ENST00000527945.1 ENST00000504230.2 |
LINC00958
|
long intergenic non-protein coding RNA 958 |
| chr7_-_87856280 | 0.15 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr9_-_116065551 | 0.15 |
ENST00000297894.5
|
RNF183
|
ring finger protein 183 |
| chr7_-_92777606 | 0.15 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr2_-_3521518 | 0.15 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chr1_-_92952433 | 0.15 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr7_-_87856303 | 0.14 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr6_-_36515177 | 0.14 |
ENST00000229812.7
|
STK38
|
serine/threonine kinase 38 |
| chr1_-_24306768 | 0.14 |
ENST00000374453.3
ENST00000453840.3 |
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr5_-_159846399 | 0.14 |
ENST00000297151.4
|
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr11_-_327537 | 0.13 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr9_-_28670283 | 0.13 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr12_-_118796910 | 0.13 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr19_+_36139125 | 0.13 |
ENST00000246554.3
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr6_+_161123270 | 0.13 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr1_-_151431647 | 0.12 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr5_+_68860949 | 0.12 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr4_-_103748880 | 0.12 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_-_116504448 | 0.12 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr2_-_220264703 | 0.12 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr19_-_44388116 | 0.11 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chr22_-_40289759 | 0.11 |
ENST00000325157.6
|
ENTHD1
|
ENTH domain containing 1 |
| chr20_-_33735070 | 0.11 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr1_+_15668240 | 0.10 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr10_+_24755416 | 0.10 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr4_-_122686261 | 0.10 |
ENST00000337677.5
|
TMEM155
|
transmembrane protein 155 |
| chr5_+_140579162 | 0.10 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chr11_+_2405833 | 0.10 |
ENST00000527343.1
ENST00000464784.2 |
CD81
|
CD81 molecule |
| chr3_+_148709128 | 0.10 |
ENST00000345003.4
ENST00000296048.6 ENST00000483267.1 |
GYG1
|
glycogenin 1 |
| chr6_+_154360357 | 0.10 |
ENST00000330432.7
ENST00000360422.4 |
OPRM1
|
opioid receptor, mu 1 |
| chr14_+_22465771 | 0.09 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr9_-_34729457 | 0.09 |
ENST00000378788.3
|
FAM205A
|
family with sequence similarity 205, member A |
| chr16_+_12059091 | 0.09 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr7_+_99202003 | 0.09 |
ENST00000609449.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr8_-_93978309 | 0.09 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr20_-_43133491 | 0.09 |
ENST00000411544.1
|
SERINC3
|
serine incorporator 3 |
| chr19_-_51920952 | 0.08 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr16_+_72088376 | 0.08 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr1_-_202897724 | 0.08 |
ENST00000435533.3
ENST00000367258.1 |
KLHL12
|
kelch-like family member 12 |
| chr20_-_29978383 | 0.08 |
ENST00000339144.3
ENST00000376321.3 |
DEFB119
|
defensin, beta 119 |
| chr6_-_87804815 | 0.07 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr4_-_74486109 | 0.07 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr8_-_93978346 | 0.07 |
ENST00000523580.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr3_+_45927994 | 0.07 |
ENST00000357632.2
ENST00000395963.2 |
CCR9
|
chemokine (C-C motif) receptor 9 |
| chr11_+_55650773 | 0.07 |
ENST00000449290.2
|
TRIM51
|
tripartite motif-containing 51 |
| chr19_-_3557570 | 0.07 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr4_-_74486347 | 0.07 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr15_+_75970672 | 0.06 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr8_-_93978333 | 0.06 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr1_-_24306798 | 0.06 |
ENST00000374452.5
ENST00000492112.2 ENST00000343255.5 ENST00000344989.6 |
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr2_-_228497888 | 0.06 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.4 | 1.8 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.4 | 1.3 | GO:0055020 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.3 | 6.8 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.3 | 5.3 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.2 | 1.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 1.9 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.2 | 2.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 0.5 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.4 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.7 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.4 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 2.9 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 1.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.8 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.2 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.2 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.9 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.3 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.4 | GO:0034627 | quinolinate biosynthetic process(GO:0019805) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.1 | 1.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.3 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.2 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.6 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 1.1 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.3 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 1.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.4 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:2000296 | regulation of hydrogen peroxide catabolic process(GO:2000295) negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.4 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 1.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.3 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |
| 0.0 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.8 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.5 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.5 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.4 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 3.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.0 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.3 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.1 | 0.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.8 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.4 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.4 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 1.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 2.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.8 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.9 | 3.4 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.5 | 1.9 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.4 | 2.0 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.4 | 1.8 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.2 | 5.3 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.4 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 1.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.3 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.1 | 2.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 1.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.8 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.9 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 1.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 5.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 2.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 1.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |