Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
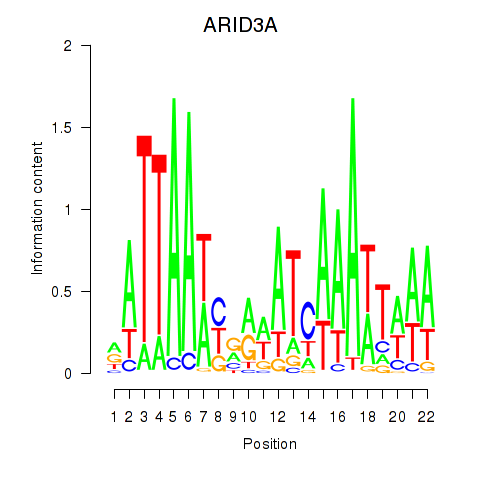
Results for ARID3A
Z-value: 0.57
Transcription factors associated with ARID3A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID3A
|
ENSG00000116017.6 | AT-rich interaction domain 3A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID3A | hg19_v2_chr19_+_926000_926046 | -0.02 | 9.1e-01 | Click! |
Activity profile of ARID3A motif
Sorted Z-values of ARID3A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_39759794 | 1.55 |
ENST00000518804.1
ENST00000519154.1 ENST00000522495.1 ENST00000522840.1 |
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr16_+_19421803 | 1.00 |
ENST00000541464.1
|
TMC5
|
transmembrane channel-like 5 |
| chr9_-_27005686 | 0.99 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr4_+_69962212 | 0.88 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr4_+_69962185 | 0.88 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr16_-_21289627 | 0.80 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr12_-_71182695 | 0.78 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr6_-_46703430 | 0.78 |
ENST00000537365.1
|
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr16_+_19422035 | 0.70 |
ENST00000381414.4
ENST00000396229.2 |
TMC5
|
transmembrane channel-like 5 |
| chr1_+_77333117 | 0.67 |
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr19_+_16059818 | 0.66 |
ENST00000322107.1
|
OR10H4
|
olfactory receptor, family 10, subfamily H, member 4 |
| chr4_-_110723194 | 0.66 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr19_+_49199209 | 0.63 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr1_-_205391178 | 0.57 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr4_-_110723335 | 0.52 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr8_+_74903580 | 0.47 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr4_+_74606223 | 0.45 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr7_+_6617039 | 0.45 |
ENST00000405731.3
ENST00000396713.2 ENST00000396707.2 ENST00000335965.6 ENST00000396709.1 ENST00000483589.1 ENST00000396706.2 |
ZDHHC4
|
zinc finger, DHHC-type containing 4 |
| chr2_+_79252834 | 0.44 |
ENST00000409471.1
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr20_+_58179582 | 0.41 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr7_-_139763521 | 0.40 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr2_+_79252822 | 0.40 |
ENST00000272324.5
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr2_+_79252804 | 0.39 |
ENST00000393897.2
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr11_+_118175132 | 0.39 |
ENST00000361763.4
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr6_+_27791862 | 0.39 |
ENST00000355057.1
|
HIST1H4J
|
histone cluster 1, H4j |
| chr6_+_47624172 | 0.39 |
ENST00000507065.1
ENST00000296862.1 |
GPR111
|
G protein-coupled receptor 111 |
| chr19_-_55458860 | 0.37 |
ENST00000592784.1
ENST00000448121.2 ENST00000340844.2 |
NLRP7
|
NLR family, pyrin domain containing 7 |
| chr5_+_140220769 | 0.37 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chr4_-_68749745 | 0.36 |
ENST00000283916.6
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr3_+_169629354 | 0.35 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr1_-_227505289 | 0.35 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr3_-_151102529 | 0.35 |
ENST00000302632.3
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12 |
| chr16_+_76587314 | 0.35 |
ENST00000563764.1
|
RP11-58C22.1
|
Uncharacterized protein |
| chr12_+_133757995 | 0.35 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chrX_+_139791917 | 0.35 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chrX_-_2882296 | 0.34 |
ENST00000438544.1
ENST00000381134.3 ENST00000545496.1 |
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1) |
| chr7_-_6048650 | 0.34 |
ENST00000382321.4
ENST00000406569.3 |
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr15_-_58306295 | 0.33 |
ENST00000559517.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr12_-_10955226 | 0.33 |
ENST00000240687.2
|
TAS2R7
|
taste receptor, type 2, member 7 |
| chr5_+_140180635 | 0.32 |
ENST00000522353.2
ENST00000532566.2 |
PCDHA3
|
protocadherin alpha 3 |
| chr19_-_55652290 | 0.32 |
ENST00000589745.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr7_-_6048702 | 0.32 |
ENST00000265849.7
|
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr10_-_28623368 | 0.31 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr17_+_71228793 | 0.31 |
ENST00000426147.2
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr18_+_61143994 | 0.31 |
ENST00000382771.4
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr1_+_207002222 | 0.30 |
ENST00000270218.6
|
IL19
|
interleukin 19 |
| chr9_-_21305312 | 0.30 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chrX_+_140982452 | 0.29 |
ENST00000544766.1
|
MAGEC3
|
melanoma antigen family C, 3 |
| chr9_-_74675521 | 0.29 |
ENST00000377024.3
|
C9orf57
|
chromosome 9 open reading frame 57 |
| chr4_-_169239921 | 0.29 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr12_-_113772835 | 0.29 |
ENST00000552014.1
ENST00000548186.1 ENST00000202831.3 ENST00000549181.1 |
SLC8B1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr7_+_156902674 | 0.28 |
ENST00000594086.1
|
AC006967.1
|
Protein LOC100996426 |
| chr10_+_124320156 | 0.28 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr9_-_69229650 | 0.27 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr9_-_21482312 | 0.27 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr7_+_138943265 | 0.27 |
ENST00000483726.1
|
UBN2
|
ubinuclein 2 |
| chr10_+_124320195 | 0.26 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr12_-_95044309 | 0.26 |
ENST00000261226.4
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr10_+_91092241 | 0.26 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr11_+_67351213 | 0.25 |
ENST00000398603.1
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr4_-_76957214 | 0.25 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr8_+_85095497 | 0.25 |
ENST00000522455.1
ENST00000521695.1 |
RALYL
|
RALY RNA binding protein-like |
| chr9_-_117420052 | 0.25 |
ENST00000423632.1
|
RP11-402G3.5
|
RP11-402G3.5 |
| chr10_+_7745303 | 0.24 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chrX_-_20159934 | 0.24 |
ENST00000379593.1
ENST00000379607.5 |
EIF1AX
|
eukaryotic translation initiation factor 1A, X-linked |
| chr8_+_85095769 | 0.24 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr3_-_101039402 | 0.24 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr1_-_31661000 | 0.23 |
ENST00000263693.1
ENST00000398657.2 ENST00000526106.1 |
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr19_-_4457776 | 0.23 |
ENST00000301281.6
|
UBXN6
|
UBX domain protein 6 |
| chr12_-_120765565 | 0.23 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr19_+_18208603 | 0.23 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr5_+_152870106 | 0.23 |
ENST00000285900.5
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1 |
| chrX_+_41193407 | 0.23 |
ENST00000457138.2
ENST00000441189.2 |
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr17_-_29641084 | 0.23 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chrX_+_41192595 | 0.22 |
ENST00000399959.2
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr7_+_89783689 | 0.22 |
ENST00000297205.2
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr11_-_86383370 | 0.22 |
ENST00000526834.1
ENST00000359636.2 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr8_-_87242589 | 0.22 |
ENST00000419776.2
ENST00000297524.3 |
SLC7A13
|
solute carrier family 7 (anionic amino acid transporter), member 13 |
| chr1_-_198990166 | 0.22 |
ENST00000427439.1
|
RP11-16L9.3
|
RP11-16L9.3 |
| chr2_+_190540711 | 0.22 |
ENST00000520309.1
|
ANKAR
|
ankyrin and armadillo repeat containing |
| chr1_-_100231349 | 0.22 |
ENST00000287474.5
ENST00000414213.1 |
FRRS1
|
ferric-chelate reductase 1 |
| chr10_+_118305435 | 0.21 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr12_+_60083118 | 0.21 |
ENST00000261187.4
ENST00000543448.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr7_+_129984630 | 0.21 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr15_-_54051831 | 0.20 |
ENST00000557913.1
ENST00000360509.5 |
WDR72
|
WD repeat domain 72 |
| chr4_+_70916119 | 0.20 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr3_-_139396853 | 0.20 |
ENST00000406164.1
ENST00000406824.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr21_+_33671264 | 0.20 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr8_+_23104130 | 0.20 |
ENST00000313219.7
ENST00000519984.1 |
CHMP7
|
charged multivesicular body protein 7 |
| chr3_-_139396801 | 0.20 |
ENST00000413939.2
ENST00000339837.5 ENST00000512391.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr16_-_84220604 | 0.20 |
ENST00000567759.1
|
TAF1C
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa |
| chr19_-_39826639 | 0.19 |
ENST00000602185.1
ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG
|
glia maturation factor, gamma |
| chr1_+_36335351 | 0.19 |
ENST00000373206.1
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr12_+_133758115 | 0.19 |
ENST00000541009.2
ENST00000592241.1 |
ZNF268
|
zinc finger protein 268 |
| chr19_+_3762645 | 0.19 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr15_-_34880646 | 0.19 |
ENST00000543376.1
|
GOLGA8A
|
golgin A8 family, member A |
| chr6_-_29324054 | 0.19 |
ENST00000543825.1
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1 |
| chr1_-_23521222 | 0.19 |
ENST00000374619.1
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr12_-_10605929 | 0.19 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr8_-_62559366 | 0.18 |
ENST00000522919.1
|
ASPH
|
aspartate beta-hydroxylase |
| chr12_-_4488872 | 0.18 |
ENST00000237837.1
|
FGF23
|
fibroblast growth factor 23 |
| chr11_-_86383650 | 0.18 |
ENST00000526944.1
ENST00000530335.1 ENST00000543262.1 ENST00000524826.1 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr3_-_139396787 | 0.18 |
ENST00000296202.7
ENST00000509291.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr10_-_69597915 | 0.18 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr17_+_7531281 | 0.18 |
ENST00000575729.1
ENST00000340624.5 |
SHBG
|
sex hormone-binding globulin |
| chrX_+_83116142 | 0.18 |
ENST00000329312.4
|
CYLC1
|
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr12_+_56114189 | 0.18 |
ENST00000548082.1
|
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr16_-_2770216 | 0.18 |
ENST00000302641.3
|
PRSS27
|
protease, serine 27 |
| chr12_+_10658201 | 0.17 |
ENST00000322446.3
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr15_-_74659978 | 0.17 |
ENST00000541301.1
ENST00000416978.1 ENST00000268053.6 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chrX_+_1387693 | 0.17 |
ENST00000381529.3
ENST00000432318.2 ENST00000361536.3 ENST00000501036.2 ENST00000381524.3 ENST00000412290.1 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr4_-_96470108 | 0.17 |
ENST00000513796.1
|
UNC5C
|
unc-5 homolog C (C. elegans) |
| chr14_+_39734482 | 0.17 |
ENST00000554392.1
ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5
|
CTAGE family, member 5 |
| chr15_-_74658493 | 0.16 |
ENST00000419019.2
ENST00000569662.1 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr16_-_84220633 | 0.16 |
ENST00000566732.1
ENST00000561955.1 ENST00000564454.1 ENST00000341690.6 ENST00000541676.1 ENST00000570117.1 ENST00000564345.1 |
TAF1C
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa |
| chr20_-_3767324 | 0.16 |
ENST00000379751.4
|
CENPB
|
centromere protein B, 80kDa |
| chr20_-_33732952 | 0.16 |
ENST00000541621.1
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr4_-_80994471 | 0.16 |
ENST00000295465.4
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr1_-_92371839 | 0.16 |
ENST00000370399.2
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr3_-_191000172 | 0.16 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr12_+_69633317 | 0.16 |
ENST00000435070.2
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr9_+_42671887 | 0.15 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr17_-_15466850 | 0.15 |
ENST00000438826.3
ENST00000225576.3 ENST00000519970.1 ENST00000518321.1 ENST00000428082.2 ENST00000522212.2 |
TVP23C
TVP23C-CDRT4
|
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) TVP23C-CDRT4 readthrough |
| chr3_+_132843652 | 0.15 |
ENST00000508711.1
|
TMEM108
|
transmembrane protein 108 |
| chr14_+_70918874 | 0.15 |
ENST00000603540.1
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr15_-_74658519 | 0.15 |
ENST00000450547.1
ENST00000358632.4 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr1_-_23520755 | 0.15 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr15_-_99057551 | 0.15 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr4_+_71263599 | 0.15 |
ENST00000399575.2
|
PROL1
|
proline rich, lacrimal 1 |
| chr2_+_190541153 | 0.15 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr9_+_90112117 | 0.15 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr9_+_79074068 | 0.14 |
ENST00000444201.2
ENST00000376730.4 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr12_+_10658489 | 0.14 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr1_-_235098935 | 0.14 |
ENST00000423175.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr3_+_36421826 | 0.14 |
ENST00000273183.3
|
STAC
|
SH3 and cysteine rich domain |
| chr18_-_13915530 | 0.14 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr2_-_190927447 | 0.14 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr1_+_211499957 | 0.14 |
ENST00000336184.2
|
TRAF5
|
TNF receptor-associated factor 5 |
| chr12_+_26126681 | 0.14 |
ENST00000542865.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr13_-_88323218 | 0.14 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr19_-_53758094 | 0.14 |
ENST00000601828.1
ENST00000598513.1 ENST00000599012.1 ENST00000333952.4 ENST00000598806.1 |
ZNF677
|
zinc finger protein 677 |
| chr6_-_32977345 | 0.14 |
ENST00000450833.2
ENST00000374813.1 ENST00000229829.5 |
HLA-DOA
|
major histocompatibility complex, class II, DO alpha |
| chr5_+_140501581 | 0.14 |
ENST00000194152.1
|
PCDHB4
|
protocadherin beta 4 |
| chr4_-_170948361 | 0.14 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr8_+_11961898 | 0.14 |
ENST00000400085.3
|
ZNF705D
|
zinc finger protein 705D |
| chr4_-_103998439 | 0.14 |
ENST00000503230.1
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr11_+_45825616 | 0.14 |
ENST00000442528.2
ENST00000456334.1 ENST00000526817.1 |
SLC35C1
|
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chrX_+_99839799 | 0.14 |
ENST00000373031.4
|
TNMD
|
tenomodulin |
| chr9_+_118916082 | 0.13 |
ENST00000328252.3
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr12_+_56114151 | 0.13 |
ENST00000547072.1
ENST00000552930.1 ENST00000257895.5 |
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chrX_-_15333736 | 0.13 |
ENST00000380470.3
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr11_+_60260248 | 0.13 |
ENST00000526784.1
ENST00000016913.4 ENST00000537076.1 ENST00000530007.1 |
MS4A12
|
membrane-spanning 4-domains, subfamily A, member 12 |
| chr12_+_113682066 | 0.13 |
ENST00000392569.4
ENST00000552542.1 |
TPCN1
|
two pore segment channel 1 |
| chrX_-_15333775 | 0.13 |
ENST00000480796.1
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr12_+_69186125 | 0.13 |
ENST00000399333.3
|
AC124890.1
|
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
| chr12_-_53601055 | 0.13 |
ENST00000552972.1
ENST00000422257.3 ENST00000267082.5 |
ITGB7
|
integrin, beta 7 |
| chr12_-_53601000 | 0.12 |
ENST00000338737.4
ENST00000549086.2 |
ITGB7
|
integrin, beta 7 |
| chr16_-_30621663 | 0.12 |
ENST00000287461.3
|
ZNF689
|
zinc finger protein 689 |
| chr21_-_37270727 | 0.12 |
ENST00000599809.1
|
FKSG68
|
FKSG68 |
| chr13_-_33780133 | 0.12 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr16_+_20499024 | 0.12 |
ENST00000593357.1
|
AC137056.1
|
Uncharacterized protein; cDNA FLJ34659 fis, clone KIDNE2018863 |
| chrX_+_75648046 | 0.12 |
ENST00000361470.2
|
MAGEE1
|
melanoma antigen family E, 1 |
| chr1_-_154600421 | 0.12 |
ENST00000368471.3
ENST00000292205.5 |
ADAR
|
adenosine deaminase, RNA-specific |
| chr12_-_122879969 | 0.12 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr10_-_115904361 | 0.11 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr12_+_51818586 | 0.11 |
ENST00000394856.1
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr12_+_51818555 | 0.11 |
ENST00000453097.2
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr15_-_65321943 | 0.11 |
ENST00000220058.4
|
MTFMT
|
mitochondrial methionyl-tRNA formyltransferase |
| chr15_-_45670924 | 0.11 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr6_-_52860171 | 0.11 |
ENST00000370963.4
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr16_+_202686 | 0.11 |
ENST00000252951.2
|
HBZ
|
hemoglobin, zeta |
| chr19_-_20748541 | 0.11 |
ENST00000427401.4
ENST00000594419.1 |
ZNF737
|
zinc finger protein 737 |
| chr20_-_29978383 | 0.11 |
ENST00000339144.3
ENST00000376321.3 |
DEFB119
|
defensin, beta 119 |
| chr1_-_72566613 | 0.11 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr10_+_7745232 | 0.10 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chrX_+_1387721 | 0.10 |
ENST00000419094.1
ENST00000381509.3 ENST00000494969.2 ENST00000355805.2 ENST00000355432.3 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr11_-_327537 | 0.10 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr16_+_30034655 | 0.10 |
ENST00000300575.2
|
C16orf92
|
chromosome 16 open reading frame 92 |
| chr22_+_24198890 | 0.10 |
ENST00000345044.6
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr19_+_37742773 | 0.10 |
ENST00000438770.2
ENST00000591116.1 ENST00000592712.1 |
AC012309.5
|
AC012309.5 |
| chr19_-_48614063 | 0.10 |
ENST00000599921.1
ENST00000599111.1 |
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent) |
| chr3_+_157827841 | 0.10 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr19_-_48614033 | 0.10 |
ENST00000354276.3
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent) |
| chr8_-_104153703 | 0.10 |
ENST00000521246.1
|
C8orf56
|
chromosome 8 open reading frame 56 |
| chr2_+_234959376 | 0.10 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr1_-_53387386 | 0.10 |
ENST00000467988.1
ENST00000358358.5 ENST00000371522.4 |
ECHDC2
|
enoyl CoA hydratase domain containing 2 |
| chrX_-_138724677 | 0.09 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr1_-_151162606 | 0.09 |
ENST00000354473.4
ENST00000368892.4 |
VPS72
|
vacuolar protein sorting 72 homolog (S. cerevisiae) |
| chr1_-_39395165 | 0.09 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr9_-_70465758 | 0.09 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr17_-_71228357 | 0.09 |
ENST00000583024.1
ENST00000403627.3 ENST00000405159.3 ENST00000581110.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr6_+_36922209 | 0.09 |
ENST00000373674.3
|
PI16
|
peptidase inhibitor 16 |
| chr5_-_43557791 | 0.09 |
ENST00000338972.4
ENST00000511321.1 ENST00000515338.1 |
PAIP1
|
poly(A) binding protein interacting protein 1 |
| chr2_-_85555385 | 0.09 |
ENST00000377386.3
|
TGOLN2
|
trans-golgi network protein 2 |
| chr2_-_109605663 | 0.09 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr8_+_104892639 | 0.09 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr19_+_52264449 | 0.09 |
ENST00000599326.1
ENST00000598953.1 |
FPR2
|
formyl peptide receptor 2 |
| chr7_-_66460563 | 0.09 |
ENST00000246868.2
|
SBDS
|
Shwachman-Bodian-Diamond syndrome |
| chr5_+_54455946 | 0.09 |
ENST00000503787.1
ENST00000296734.6 ENST00000515370.1 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr10_-_100995603 | 0.09 |
ENST00000370552.3
ENST00000370549.1 |
HPSE2
|
heparanase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID3A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.4 | 1.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 0.5 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.8 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.1 | 0.6 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.8 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.4 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 1.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.5 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.4 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 0.3 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 0.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.2 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.2 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 0.4 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 0.6 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.4 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.2 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.7 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.3 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.3 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.2 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.3 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.3 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.0 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.1 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 0.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |
| 0.0 | 1.0 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.2 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.0 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.3 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 0.7 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 0.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 0.2 | GO:0043159 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.0 | 0.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.2 | 0.8 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.2 | 0.5 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.1 | 0.6 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 1.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.7 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.1 | 1.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.4 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.2 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 1.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.3 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 0.1 | 0.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.5 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 1.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.6 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.4 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.3 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.0 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |