Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
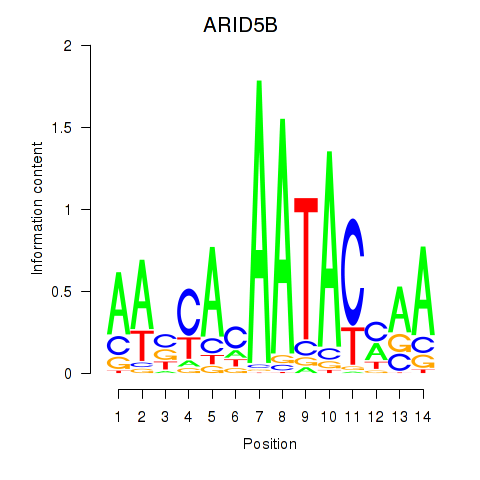
Results for ARID5B
Z-value: 1.00
Transcription factors associated with ARID5B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5B
|
ENSG00000150347.10 | AT-rich interaction domain 5B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5B | hg19_v2_chr10_+_63661053_63661079 | -0.10 | 6.1e-01 | Click! |
Activity profile of ARID5B motif
Sorted Z-values of ARID5B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_153085984 | 4.81 |
ENST00000468739.1
|
SPRR2F
|
small proline-rich protein 2F |
| chr1_-_205391178 | 4.56 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr1_-_153066998 | 4.54 |
ENST00000368750.3
|
SPRR2E
|
small proline-rich protein 2E |
| chr1_-_149459549 | 4.13 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr5_+_154393260 | 3.90 |
ENST00000435029.4
|
KIF4B
|
kinesin family member 4B |
| chr1_+_152975488 | 3.64 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr15_-_80263506 | 3.53 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr4_-_57524061 | 3.21 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chr4_-_159094194 | 3.16 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr22_+_19467261 | 3.12 |
ENST00000455750.1
ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45
|
cell division cycle 45 |
| chr5_+_68462837 | 2.91 |
ENST00000256442.5
|
CCNB1
|
cyclin B1 |
| chr21_+_30502806 | 2.89 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr7_-_24797546 | 2.77 |
ENST00000414428.1
ENST00000419307.1 ENST00000342947.3 |
DFNA5
|
deafness, autosomal dominant 5 |
| chr12_-_8815215 | 2.67 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr12_-_8815299 | 2.58 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr12_-_8814669 | 2.38 |
ENST00000535411.1
ENST00000540087.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr3_+_99357319 | 2.36 |
ENST00000452013.1
ENST00000261037.3 ENST00000273342.4 |
COL8A1
|
collagen, type VIII, alpha 1 |
| chr22_-_29107919 | 2.27 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr12_-_10007448 | 2.27 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr12_+_51236703 | 2.07 |
ENST00000551456.1
ENST00000398458.3 |
TMPRSS12
|
transmembrane (C-terminal) protease, serine 12 |
| chr3_-_74570291 | 2.00 |
ENST00000263665.6
|
CNTN3
|
contactin 3 (plasmacytoma associated) |
| chr6_+_126661253 | 2.00 |
ENST00000368326.1
ENST00000368325.1 ENST00000368328.4 |
CENPW
|
centromere protein W |
| chr1_-_76076793 | 1.91 |
ENST00000370859.3
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr14_-_69263043 | 1.86 |
ENST00000408913.2
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr17_+_61086917 | 1.79 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr1_-_168106536 | 1.68 |
ENST00000537209.1
ENST00000361697.2 ENST00000546300.1 ENST00000367835.1 |
GPR161
|
G protein-coupled receptor 161 |
| chr7_-_56119238 | 1.60 |
ENST00000275605.3
ENST00000395471.3 |
PSPH
|
phosphoserine phosphatase |
| chr11_-_121986923 | 1.45 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chrX_+_82763265 | 1.42 |
ENST00000373200.2
|
POU3F4
|
POU class 3 homeobox 4 |
| chr9_+_706842 | 1.39 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr3_-_185538849 | 1.39 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr15_-_91565743 | 1.36 |
ENST00000535843.1
|
VPS33B
|
vacuolar protein sorting 33 homolog B (yeast) |
| chr17_-_36358166 | 1.35 |
ENST00000537432.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr1_-_43855444 | 1.31 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr6_+_24775153 | 1.30 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr14_+_32798547 | 1.29 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr10_-_75193308 | 1.28 |
ENST00000299432.2
|
MSS51
|
MSS51 mitochondrial translational activator |
| chr2_-_40680578 | 1.28 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr3_-_197024394 | 1.27 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chrX_+_102585124 | 1.22 |
ENST00000332431.4
ENST00000372666.1 |
TCEAL7
|
transcription elongation factor A (SII)-like 7 |
| chr12_-_57146095 | 1.21 |
ENST00000550770.1
ENST00000338193.6 |
PRIM1
|
primase, DNA, polypeptide 1 (49kDa) |
| chr1_+_32608566 | 1.15 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr14_+_32798462 | 1.14 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr10_-_29923893 | 1.14 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr15_-_91565770 | 1.10 |
ENST00000535906.1
ENST00000333371.3 |
VPS33B
|
vacuolar protein sorting 33 homolog B (yeast) |
| chr1_+_78245303 | 1.10 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr7_-_56119156 | 1.09 |
ENST00000421312.1
ENST00000416592.1 |
PSPH
|
phosphoserine phosphatase |
| chr12_+_56324933 | 1.08 |
ENST00000549629.1
ENST00000555218.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr4_-_80247162 | 1.07 |
ENST00000286794.4
|
NAA11
|
N(alpha)-acetyltransferase 11, NatA catalytic subunit |
| chr3_+_190333097 | 1.06 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr17_-_64225508 | 1.04 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr4_+_106631966 | 1.01 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr2_-_89327228 | 0.99 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11 |
| chr11_+_73882144 | 0.97 |
ENST00000328257.8
|
PPME1
|
protein phosphatase methylesterase 1 |
| chr11_+_18417813 | 0.89 |
ENST00000540430.1
ENST00000379412.5 |
LDHA
|
lactate dehydrogenase A |
| chr1_-_85742773 | 0.87 |
ENST00000370580.1
|
BCL10
|
B-cell CLL/lymphoma 10 |
| chr11_+_73882311 | 0.83 |
ENST00000398427.4
ENST00000544401.1 |
PPME1
|
protein phosphatase methylesterase 1 |
| chr5_+_135496675 | 0.81 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr3_+_102153859 | 0.79 |
ENST00000306176.1
ENST00000466937.1 |
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr3_+_63428752 | 0.79 |
ENST00000295894.5
|
SYNPR
|
synaptoporin |
| chr11_-_78052923 | 0.78 |
ENST00000340149.2
|
GAB2
|
GRB2-associated binding protein 2 |
| chr10_-_69834973 | 0.76 |
ENST00000395187.2
|
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr1_-_76076759 | 0.76 |
ENST00000370855.5
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr2_+_133874577 | 0.75 |
ENST00000596384.1
|
AC011755.1
|
HCG2006742; Protein LOC100996685 |
| chr6_+_42896865 | 0.75 |
ENST00000372836.4
ENST00000394142.3 |
CNPY3
|
canopy FGF signaling regulator 3 |
| chr12_+_56324756 | 0.74 |
ENST00000331886.5
ENST00000555090.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr10_-_69835099 | 0.74 |
ENST00000373700.4
|
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr21_+_40177143 | 0.73 |
ENST00000360214.3
|
ETS2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr18_+_616711 | 0.72 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr2_-_231825668 | 0.72 |
ENST00000392039.2
|
GPR55
|
G protein-coupled receptor 55 |
| chr4_+_147096837 | 0.71 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr3_-_98241358 | 0.71 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr2_+_242498135 | 0.69 |
ENST00000318407.3
|
BOK
|
BCL2-related ovarian killer |
| chr15_-_55563072 | 0.69 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_+_79258547 | 0.67 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr6_-_137494775 | 0.65 |
ENST00000349184.4
ENST00000296980.2 ENST00000339602.3 |
IL22RA2
|
interleukin 22 receptor, alpha 2 |
| chr3_-_113897899 | 0.65 |
ENST00000383673.2
ENST00000295881.7 |
DRD3
|
dopamine receptor D3 |
| chr3_+_68053359 | 0.64 |
ENST00000478136.1
|
FAM19A1
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
| chr19_-_56826157 | 0.63 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr3_-_98241760 | 0.61 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr4_-_71532339 | 0.61 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr18_-_51107372 | 0.61 |
ENST00000565170.1
|
RP11-202D1.2
|
RP11-202D1.2 |
| chr8_-_124054362 | 0.60 |
ENST00000405944.3
|
DERL1
|
derlin 1 |
| chr8_+_55528627 | 0.60 |
ENST00000220676.1
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr10_+_120116527 | 0.60 |
ENST00000445161.1
|
LINC00867
|
long intergenic non-protein coding RNA 867 |
| chr12_+_79258444 | 0.59 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr10_-_69835001 | 0.59 |
ENST00000513996.1
ENST00000412272.2 ENST00000395198.3 ENST00000492996.2 |
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr4_+_71588372 | 0.55 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr4_+_187187098 | 0.54 |
ENST00000403665.2
ENST00000264692.4 |
F11
|
coagulation factor XI |
| chr13_+_53216565 | 0.52 |
ENST00000357495.2
|
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2 |
| chr6_-_30080863 | 0.51 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr1_+_87458692 | 0.50 |
ENST00000370548.2
ENST00000356813.4 |
RP5-1052I5.2
HS2ST1
|
Heparan sulfate 2-O-sulfotransferase 1 heparan sulfate 2-O-sulfotransferase 1 |
| chr5_-_179047881 | 0.50 |
ENST00000521173.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr12_+_57146233 | 0.48 |
ENST00000554643.1
ENST00000556650.1 ENST00000554150.1 ENST00000554155.1 |
HSD17B6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr8_+_27632047 | 0.48 |
ENST00000397418.2
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr11_+_30253410 | 0.48 |
ENST00000533718.1
|
FSHB
|
follicle stimulating hormone, beta polypeptide |
| chr3_+_111805182 | 0.48 |
ENST00000430855.1
ENST00000431717.2 ENST00000264848.5 |
C3orf52
|
chromosome 3 open reading frame 52 |
| chr2_+_89999259 | 0.47 |
ENST00000558026.1
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr9_+_77230499 | 0.47 |
ENST00000396204.2
|
RORB
|
RAR-related orphan receptor B |
| chr9_-_32552551 | 0.47 |
ENST00000360538.2
ENST00000379858.1 |
TOPORS
|
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase |
| chr1_+_86934526 | 0.47 |
ENST00000394711.1
|
CLCA1
|
chloride channel accessory 1 |
| chr2_+_197577841 | 0.45 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr6_+_29068386 | 0.44 |
ENST00000377171.3
|
OR2J1
|
olfactory receptor, family 2, subfamily J, member 1 (gene/pseudogene) |
| chr17_+_37356555 | 0.44 |
ENST00000579374.1
|
RPL19
|
ribosomal protein L19 |
| chr4_+_95376396 | 0.44 |
ENST00000508216.1
ENST00000514743.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr12_+_8309630 | 0.44 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr8_+_66933779 | 0.43 |
ENST00000276570.5
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr19_+_17326141 | 0.42 |
ENST00000445667.2
ENST00000263897.5 |
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr17_+_37356586 | 0.42 |
ENST00000579260.1
ENST00000582193.1 |
RPL19
|
ribosomal protein L19 |
| chr17_+_37356528 | 0.41 |
ENST00000225430.4
|
RPL19
|
ribosomal protein L19 |
| chr15_+_69857515 | 0.40 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr12_+_93096619 | 0.39 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr3_+_183770835 | 0.39 |
ENST00000318351.1
|
HTR3C
|
5-hydroxytryptamine (serotonin) receptor 3C, ionotropic |
| chr12_+_128399965 | 0.38 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr6_-_138893661 | 0.38 |
ENST00000427025.2
|
NHSL1
|
NHS-like 1 |
| chr19_+_35849362 | 0.38 |
ENST00000327809.4
|
FFAR3
|
free fatty acid receptor 3 |
| chr2_-_178753465 | 0.37 |
ENST00000389683.3
|
PDE11A
|
phosphodiesterase 11A |
| chr8_-_66474884 | 0.37 |
ENST00000520902.1
|
CTD-3025N20.2
|
CTD-3025N20.2 |
| chr7_+_153749732 | 0.37 |
ENST00000377770.3
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr2_+_223725652 | 0.36 |
ENST00000357430.3
ENST00000392066.3 |
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr17_-_10452929 | 0.36 |
ENST00000532183.2
ENST00000397183.2 ENST00000420805.1 |
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult |
| chr14_-_65409502 | 0.36 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr7_-_115608304 | 0.36 |
ENST00000457268.1
|
TFEC
|
transcription factor EC |
| chr4_-_77328458 | 0.36 |
ENST00000388914.3
ENST00000434846.2 |
CCDC158
|
coiled-coil domain containing 158 |
| chr9_+_35806082 | 0.35 |
ENST00000447210.1
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chrX_+_106769876 | 0.35 |
ENST00000439554.1
|
FRMPD3
|
FERM and PDZ domain containing 3 |
| chr9_+_116267536 | 0.34 |
ENST00000374136.1
|
RGS3
|
regulator of G-protein signaling 3 |
| chr17_+_3379284 | 0.34 |
ENST00000263080.2
|
ASPA
|
aspartoacylase |
| chr17_+_60447579 | 0.34 |
ENST00000450662.2
|
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr18_+_21032781 | 0.34 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr11_-_104769141 | 0.34 |
ENST00000508062.1
ENST00000422698.2 |
CASP12
|
caspase 12 (gene/pseudogene) |
| chr3_+_184018352 | 0.34 |
ENST00000435761.1
ENST00000439383.1 |
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr15_+_36994210 | 0.34 |
ENST00000562489.1
|
C15orf41
|
chromosome 15 open reading frame 41 |
| chr15_+_80364901 | 0.33 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr3_-_113897545 | 0.33 |
ENST00000467632.1
|
DRD3
|
dopamine receptor D3 |
| chrX_-_11308598 | 0.31 |
ENST00000380717.3
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr5_+_161495038 | 0.30 |
ENST00000393933.4
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr12_+_54366894 | 0.30 |
ENST00000546378.1
ENST00000243082.4 |
HOXC11
|
homeobox C11 |
| chr11_+_46193466 | 0.30 |
ENST00000533793.1
|
RP11-702F3.3
|
RP11-702F3.3 |
| chr12_-_56321397 | 0.30 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr5_-_150138551 | 0.29 |
ENST00000446090.2
ENST00000447998.2 |
DCTN4
|
dynactin 4 (p62) |
| chr17_-_3599696 | 0.29 |
ENST00000225328.5
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr1_+_150266256 | 0.29 |
ENST00000309092.7
ENST00000369084.5 |
MRPS21
|
mitochondrial ribosomal protein S21 |
| chr15_+_51669444 | 0.29 |
ENST00000396399.2
|
GLDN
|
gliomedin |
| chr2_-_74619152 | 0.28 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chrX_+_86772787 | 0.28 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chr17_-_3599492 | 0.27 |
ENST00000435558.1
ENST00000345901.3 |
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr14_-_24711865 | 0.27 |
ENST00000399423.4
ENST00000267415.7 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr12_-_91539918 | 0.27 |
ENST00000548218.1
|
DCN
|
decorin |
| chr16_+_58426296 | 0.26 |
ENST00000426538.2
ENST00000328514.7 ENST00000318129.5 |
GINS3
|
GINS complex subunit 3 (Psf3 homolog) |
| chr14_-_24711806 | 0.26 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr5_+_137225158 | 0.25 |
ENST00000290431.5
|
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr11_-_111794446 | 0.25 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr2_+_168675182 | 0.25 |
ENST00000305861.1
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr9_-_21202204 | 0.25 |
ENST00000239347.3
|
IFNA7
|
interferon, alpha 7 |
| chr5_+_137225125 | 0.24 |
ENST00000350250.4
ENST00000508638.1 ENST00000502810.1 ENST00000508883.1 |
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr2_+_119699864 | 0.24 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr1_-_114414316 | 0.23 |
ENST00000528414.1
ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr3_+_196594727 | 0.23 |
ENST00000445299.2
ENST00000323460.5 ENST00000419026.1 |
SENP5
|
SUMO1/sentrin specific peptidase 5 |
| chr3_+_130150307 | 0.23 |
ENST00000512836.1
|
COL6A5
|
collagen, type VI, alpha 5 |
| chr9_+_114287433 | 0.23 |
ENST00000358151.4
ENST00000355824.3 ENST00000374374.3 ENST00000309235.5 |
ZNF483
|
zinc finger protein 483 |
| chr4_-_69817481 | 0.22 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr1_+_186798073 | 0.22 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr4_-_90229142 | 0.21 |
ENST00000609438.1
|
GPRIN3
|
GPRIN family member 3 |
| chr2_-_74618964 | 0.20 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr12_-_10962767 | 0.20 |
ENST00000240691.2
|
TAS2R9
|
taste receptor, type 2, member 9 |
| chr5_+_92919043 | 0.20 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr3_+_178276488 | 0.20 |
ENST00000432997.1
ENST00000455865.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr12_+_56390964 | 0.19 |
ENST00000356124.4
ENST00000266971.3 ENST00000394115.2 ENST00000547586.1 ENST00000552258.1 ENST00000548274.1 ENST00000546833.1 |
SUOX
|
sulfite oxidase |
| chr1_+_151739131 | 0.19 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr12_-_56321649 | 0.19 |
ENST00000454792.2
ENST00000408946.2 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr4_+_187187337 | 0.17 |
ENST00000492972.2
|
F11
|
coagulation factor XI |
| chr1_+_3773825 | 0.16 |
ENST00000378209.3
ENST00000338895.3 ENST00000378212.2 ENST00000341385.3 |
DFFB
|
DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) |
| chr12_-_68553512 | 0.16 |
ENST00000229135.3
|
IFNG
|
interferon, gamma |
| chr9_+_71736177 | 0.16 |
ENST00000606364.1
ENST00000453658.2 |
TJP2
|
tight junction protein 2 |
| chr1_-_31538517 | 0.16 |
ENST00000440538.2
ENST00000423018.2 ENST00000424085.2 ENST00000426105.2 ENST00000257075.5 ENST00000373747.3 ENST00000525843.1 ENST00000373742.2 |
PUM1
|
pumilio RNA-binding family member 1 |
| chr1_+_92632542 | 0.16 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr4_+_184020398 | 0.15 |
ENST00000403733.3
ENST00000378925.3 |
WWC2
|
WW and C2 domain containing 2 |
| chr2_-_158345462 | 0.13 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chrX_-_101410762 | 0.13 |
ENST00000543160.1
ENST00000333643.3 |
BEX5
|
brain expressed, X-linked 5 |
| chr4_+_76871883 | 0.12 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr1_+_150039369 | 0.11 |
ENST00000369130.3
ENST00000369128.5 |
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr12_+_128399917 | 0.11 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr18_-_25616519 | 0.11 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr2_-_163695238 | 0.11 |
ENST00000328032.4
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr8_-_143957213 | 0.10 |
ENST00000519285.1
|
CYP11B1
|
cytochrome P450, family 11, subfamily B, polypeptide 1 |
| chr5_-_64920115 | 0.09 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr2_+_181845843 | 0.08 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr5_+_131993856 | 0.08 |
ENST00000304506.3
|
IL13
|
interleukin 13 |
| chr12_-_67197760 | 0.08 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr10_+_126150369 | 0.08 |
ENST00000392757.4
ENST00000368842.5 ENST00000368839.1 |
LHPP
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr6_+_25754927 | 0.08 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr12_+_52282001 | 0.07 |
ENST00000340970.4
|
ANKRD33
|
ankyrin repeat domain 33 |
| chr17_-_21454898 | 0.07 |
ENST00000391411.5
ENST00000412778.3 |
C17orf51
|
chromosome 17 open reading frame 51 |
| chr4_-_89978299 | 0.07 |
ENST00000511976.1
ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr1_-_115292591 | 0.07 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr2_-_214016314 | 0.07 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr5_-_98262240 | 0.06 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chrX_+_140084756 | 0.06 |
ENST00000449283.1
|
SPANXB2
|
SPANX family, member B2 |
| chr2_-_114300213 | 0.05 |
ENST00000446595.1
ENST00000416105.1 ENST00000450636.1 ENST00000416758.1 |
RP11-395L14.4
|
RP11-395L14.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0031938 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 1.0 | 2.9 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.8 | 2.3 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.6 | 1.9 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.5 | 1.4 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.4 | 1.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.4 | 2.5 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.4 | 2.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.3 | 1.3 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.3 | 0.9 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.3 | 1.1 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.2 | 3.9 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 1.0 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.2 | 1.3 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.2 | 2.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 1.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 1.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 13.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 0.7 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.2 | 7.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.2 | 0.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.2 | 2.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.7 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 1.5 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 0.8 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 3.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 1.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 2.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.7 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.4 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.3 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 0.7 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 3.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.7 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.6 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 0.5 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.1 | 0.4 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 1.0 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 1.3 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 | 0.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.5 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.1 | 0.3 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.5 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.2 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 1.8 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.5 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.7 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.1 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.9 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.9 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 1.8 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 2.4 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 2.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 2.8 | GO:0060113 | inner ear receptor cell differentiation(GO:0060113) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 1.3 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 1.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.3 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 1.3 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.7 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 1.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.3 | GO:0006625 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.6 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.0 | 1.4 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 0.3 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.5 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.6 | 2.9 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.3 | 7.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 2.4 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.3 | 1.3 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.2 | 2.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 13.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 2.5 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.2 | 0.9 | GO:0046696 | CBM complex(GO:0032449) lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 1.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 0.6 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.1 | 1.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.3 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.5 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 0.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 3.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 1.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.6 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 1.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 4.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.4 | 2.9 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 1.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 4.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 1.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.3 | 2.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 1.1 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.2 | 1.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 4.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 3.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 1.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.2 | 1.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 1.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.7 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 2.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 1.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 3.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 0.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 2.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 2.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 7.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.8 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.6 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.5 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.9 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 1.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.6 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.7 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.5 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 3.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 7.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 2.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 4.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 3.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 3.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 1.9 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 1.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 2.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.9 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.9 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.9 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |