Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
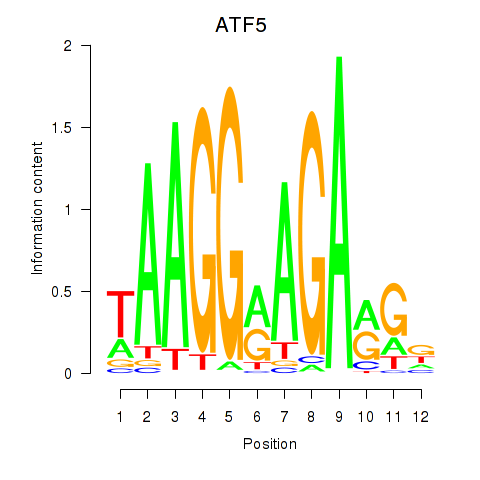
Results for ATF5
Z-value: 0.82
Transcription factors associated with ATF5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF5
|
ENSG00000169136.4 | activating transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF5 | hg19_v2_chr19_+_50431959_50431998 | -0.34 | 6.2e-02 | Click! |
Activity profile of ATF5 motif
Sorted Z-values of ATF5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_41740181 | 3.47 |
ENST00000442711.1
|
INHBA
|
inhibin, beta A |
| chr5_-_39219705 | 3.37 |
ENST00000351578.6
|
FYB
|
FYN binding protein |
| chr1_-_17304771 | 3.01 |
ENST00000375534.3
|
MFAP2
|
microfibrillar-associated protein 2 |
| chr5_-_39219641 | 2.49 |
ENST00000509072.1
ENST00000504542.1 ENST00000505428.1 ENST00000506557.1 |
FYB
|
FYN binding protein |
| chr2_-_113542063 | 2.31 |
ENST00000263339.3
|
IL1A
|
interleukin 1, alpha |
| chr5_+_156887027 | 2.03 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chr1_+_209859510 | 1.77 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr6_+_43738444 | 1.60 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr12_-_25101920 | 1.53 |
ENST00000539780.1
ENST00000546285.1 ENST00000342945.5 |
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr5_+_154393260 | 1.46 |
ENST00000435029.4
|
KIF4B
|
kinesin family member 4B |
| chr2_-_26205340 | 1.41 |
ENST00000264712.3
|
KIF3C
|
kinesin family member 3C |
| chr15_+_80733570 | 1.29 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr19_-_44285401 | 1.22 |
ENST00000262888.3
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr1_+_16083154 | 1.19 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr11_+_124735282 | 1.18 |
ENST00000397801.1
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr20_+_58179582 | 1.18 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr16_-_72127456 | 1.07 |
ENST00000562153.1
|
TXNL4B
|
thioredoxin-like 4B |
| chr7_-_3083573 | 0.96 |
ENST00000396946.4
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr3_+_101504200 | 0.86 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr6_-_88411911 | 0.86 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr4_+_77172847 | 0.79 |
ENST00000515604.1
ENST00000539752.1 ENST00000424749.2 |
FAM47E
FAM47E-STBD1
FAM47E
|
uncharacterized protein LOC100631383 FAM47E-STBD1 readthrough family with sequence similarity 47, member E |
| chr17_+_33914460 | 0.78 |
ENST00000537622.2
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr5_+_137203465 | 0.78 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr11_-_11747257 | 0.77 |
ENST00000601641.1
|
AC131935.1
|
AC131935.1 |
| chr1_+_206579736 | 0.74 |
ENST00000439126.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr14_-_36988882 | 0.72 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr2_+_234601512 | 0.71 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr15_+_63481668 | 0.71 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr17_+_33914276 | 0.71 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr7_+_150688166 | 0.70 |
ENST00000461406.1
|
NOS3
|
nitric oxide synthase 3 (endothelial cell) |
| chr4_-_159956333 | 0.69 |
ENST00000434826.2
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr14_+_50065459 | 0.68 |
ENST00000318317.4
|
LRR1
|
leucine rich repeat protein 1 |
| chrX_+_131157609 | 0.65 |
ENST00000496850.1
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr12_-_72057638 | 0.65 |
ENST00000552037.1
ENST00000378743.3 |
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr8_-_131028869 | 0.62 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr12_+_56915776 | 0.61 |
ENST00000550726.1
ENST00000542360.1 |
RBMS2
|
RNA binding motif, single stranded interacting protein 2 |
| chr2_-_188419078 | 0.61 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr7_+_73498118 | 0.61 |
ENST00000336180.2
|
LIMK1
|
LIM domain kinase 1 |
| chr12_+_125811162 | 0.60 |
ENST00000299308.3
|
TMEM132B
|
transmembrane protein 132B |
| chr3_-_99569821 | 0.59 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr12_+_32655048 | 0.58 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr14_+_23845995 | 0.58 |
ENST00000359320.3
|
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr12_-_56843161 | 0.57 |
ENST00000554616.1
ENST00000553532.1 ENST00000229201.4 |
TIMELESS
|
timeless circadian clock |
| chr2_-_190927447 | 0.57 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr14_+_100111447 | 0.57 |
ENST00000330710.5
ENST00000357223.2 |
HHIPL1
|
HHIP-like 1 |
| chr2_-_188419200 | 0.57 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr14_+_23846210 | 0.57 |
ENST00000339180.4
ENST00000342473.4 ENST00000397227.3 ENST00000555731.1 |
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr20_+_62492566 | 0.56 |
ENST00000369916.3
|
ABHD16B
|
abhydrolase domain containing 16B |
| chr19_+_48969094 | 0.55 |
ENST00000595676.1
|
CTC-273B12.7
|
Uncharacterized protein |
| chr14_+_50065376 | 0.55 |
ENST00000298288.6
|
LRR1
|
leucine rich repeat protein 1 |
| chr2_-_87018784 | 0.54 |
ENST00000283635.3
ENST00000538832.1 |
CD8A
|
CD8a molecule |
| chr11_+_57310114 | 0.54 |
ENST00000527972.1
ENST00000399154.2 |
SMTNL1
|
smoothelin-like 1 |
| chr3_-_47950745 | 0.53 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr5_+_34757309 | 0.52 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr8_+_38677850 | 0.52 |
ENST00000518809.1
ENST00000520611.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr22_-_36236623 | 0.51 |
ENST00000405409.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr4_+_159443090 | 0.50 |
ENST00000343542.5
ENST00000470033.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr22_-_36357671 | 0.49 |
ENST00000408983.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr12_+_56915713 | 0.48 |
ENST00000262031.5
ENST00000552247.2 |
RBMS2
|
RNA binding motif, single stranded interacting protein 2 |
| chr6_+_29079668 | 0.48 |
ENST00000377169.1
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3 |
| chr4_+_159442878 | 0.48 |
ENST00000307765.5
ENST00000423548.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr19_-_38916802 | 0.46 |
ENST00000587738.1
|
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr7_+_128399002 | 0.45 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr2_-_87017985 | 0.44 |
ENST00000352580.3
|
CD8A
|
CD8a molecule |
| chr8_-_95449155 | 0.43 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chrX_+_122318318 | 0.43 |
ENST00000371251.1
ENST00000371256.5 |
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chrX_+_122318006 | 0.43 |
ENST00000371266.1
ENST00000264357.5 |
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chrX_+_122318224 | 0.42 |
ENST00000542149.1
|
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr9_+_116917807 | 0.42 |
ENST00000356083.3
|
COL27A1
|
collagen, type XXVII, alpha 1 |
| chr12_-_54778471 | 0.41 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr12_+_10124001 | 0.40 |
ENST00000396507.3
ENST00000304361.4 ENST00000434319.2 |
CLEC12A
|
C-type lectin domain family 12, member A |
| chr15_-_75871589 | 0.39 |
ENST00000306726.2
|
PTPN9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr18_-_24283586 | 0.39 |
ENST00000579458.1
|
U3
|
Small nucleolar RNA U3 |
| chr12_-_16761007 | 0.39 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr14_+_70918874 | 0.38 |
ENST00000603540.1
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr5_+_137203541 | 0.38 |
ENST00000421631.2
|
MYOT
|
myotilin |
| chr16_+_28943260 | 0.38 |
ENST00000538922.1
ENST00000324662.3 ENST00000567541.1 |
CD19
|
CD19 molecule |
| chr5_+_137203557 | 0.38 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chr16_+_72127670 | 0.37 |
ENST00000536867.1
|
DHX38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr9_+_2621798 | 0.37 |
ENST00000382100.3
|
VLDLR
|
very low density lipoprotein receptor |
| chr16_+_72127461 | 0.36 |
ENST00000268482.3
ENST00000566794.1 |
DHX38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr8_-_100025238 | 0.36 |
ENST00000521696.1
|
RP11-410L14.2
|
RP11-410L14.2 |
| chr1_-_27930102 | 0.35 |
ENST00000247087.5
ENST00000374011.2 |
AHDC1
|
AT hook, DNA binding motif, containing 1 |
| chr3_+_38206975 | 0.35 |
ENST00000446845.1
ENST00000311806.3 |
OXSR1
|
oxidative stress responsive 1 |
| chr10_+_102505468 | 0.34 |
ENST00000361791.3
ENST00000355243.3 ENST00000428433.1 ENST00000370296.2 |
PAX2
|
paired box 2 |
| chr2_-_208994548 | 0.34 |
ENST00000282141.3
|
CRYGC
|
crystallin, gamma C |
| chr17_-_73149921 | 0.34 |
ENST00000481647.1
ENST00000470924.1 |
HN1
|
hematological and neurological expressed 1 |
| chr12_-_1920886 | 0.34 |
ENST00000536846.2
ENST00000538027.2 ENST00000538450.1 |
CACNA2D4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr4_+_159443024 | 0.34 |
ENST00000448688.2
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr5_+_82767487 | 0.33 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chr6_-_132967142 | 0.32 |
ENST00000275216.1
|
TAAR1
|
trace amine associated receptor 1 |
| chr5_+_175223313 | 0.32 |
ENST00000359546.4
|
CPLX2
|
complexin 2 |
| chr6_+_114178512 | 0.32 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr5_+_82767284 | 0.31 |
ENST00000265077.3
|
VCAN
|
versican |
| chr2_-_209119831 | 0.30 |
ENST00000345146.2
|
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr9_+_2622085 | 0.30 |
ENST00000382099.2
|
VLDLR
|
very low density lipoprotein receptor |
| chr8_+_22462532 | 0.30 |
ENST00000389279.3
|
CCAR2
|
cell cycle and apoptosis regulator 2 |
| chr22_-_36236265 | 0.30 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr19_-_38916839 | 0.30 |
ENST00000433821.2
ENST00000426920.2 ENST00000587753.1 ENST00000454404.2 ENST00000293062.9 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr1_+_66999799 | 0.29 |
ENST00000371035.3
ENST00000371036.3 ENST00000371037.4 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr9_-_71155783 | 0.29 |
ENST00000377311.3
|
TMEM252
|
transmembrane protein 252 |
| chr16_-_72128270 | 0.29 |
ENST00000426362.2
|
TXNL4B
|
thioredoxin-like 4B |
| chr2_-_209118974 | 0.29 |
ENST00000415913.1
ENST00000415282.1 ENST00000446179.1 |
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr8_-_623547 | 0.29 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chr9_-_38069208 | 0.28 |
ENST00000377707.3
ENST00000377700.4 |
SHB
|
Src homology 2 domain containing adaptor protein B |
| chr16_-_72127550 | 0.27 |
ENST00000268483.3
|
TXNL4B
|
thioredoxin-like 4B |
| chr22_-_37215523 | 0.27 |
ENST00000216200.5
|
PVALB
|
parvalbumin |
| chr5_+_82767583 | 0.27 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chr20_+_54987305 | 0.27 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr20_+_54987168 | 0.27 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chrX_+_122318113 | 0.27 |
ENST00000371264.3
|
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr12_-_115121962 | 0.27 |
ENST00000349155.2
|
TBX3
|
T-box 3 |
| chr2_-_128784846 | 0.26 |
ENST00000259235.3
ENST00000357702.5 ENST00000424298.1 |
SAP130
|
Sin3A-associated protein, 130kDa |
| chr7_-_115670792 | 0.26 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr19_+_15160130 | 0.26 |
ENST00000427043.3
|
CASP14
|
caspase 14, apoptosis-related cysteine peptidase |
| chr19_-_38916822 | 0.26 |
ENST00000586305.1
|
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr3_-_62860878 | 0.26 |
ENST00000283269.9
|
CADPS
|
Ca++-dependent secretion activator |
| chr16_+_30075463 | 0.25 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr8_+_22462145 | 0.25 |
ENST00000308511.4
ENST00000523801.1 ENST00000521301.1 |
CCAR2
|
cell cycle and apoptosis regulator 2 |
| chr2_-_145278475 | 0.25 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_-_52967600 | 0.25 |
ENST00000549343.1
ENST00000305620.2 |
KRT74
|
keratin 74 |
| chr1_+_151739131 | 0.25 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr7_-_115670804 | 0.24 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr2_+_153191706 | 0.24 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr18_-_19748379 | 0.24 |
ENST00000579431.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr3_-_62861012 | 0.23 |
ENST00000357948.3
ENST00000383710.4 |
CADPS
|
Ca++-dependent secretion activator |
| chr5_+_92919043 | 0.23 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr9_-_23825956 | 0.23 |
ENST00000397312.2
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr5_-_134788086 | 0.23 |
ENST00000537858.1
|
TIFAB
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chr12_+_120884222 | 0.22 |
ENST00000551765.1
ENST00000229384.5 |
GATC
|
glutamyl-tRNA(Gln) amidotransferase, subunit C |
| chr7_+_132937820 | 0.21 |
ENST00000393161.2
ENST00000253861.4 |
EXOC4
|
exocyst complex component 4 |
| chr1_-_67896009 | 0.20 |
ENST00000370990.5
|
SERBP1
|
SERPINE1 mRNA binding protein 1 |
| chr10_+_135207623 | 0.20 |
ENST00000317502.6
ENST00000432508.3 |
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr18_-_812517 | 0.19 |
ENST00000584307.1
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr18_-_812231 | 0.19 |
ENST00000314574.4
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr9_-_23826298 | 0.19 |
ENST00000380117.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr2_-_100759037 | 0.19 |
ENST00000317233.4
ENST00000423966.1 ENST00000416492.1 |
AFF3
|
AF4/FMR2 family, member 3 |
| chr3_-_62860704 | 0.18 |
ENST00000490353.2
|
CADPS
|
Ca++-dependent secretion activator |
| chr12_-_49351303 | 0.18 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr5_+_79615790 | 0.18 |
ENST00000296739.4
|
SPZ1
|
spermatogenic leucine zipper 1 |
| chr1_-_22109682 | 0.18 |
ENST00000400301.1
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr22_-_26875345 | 0.18 |
ENST00000398141.1
|
HPS4
|
Hermansky-Pudlak syndrome 4 |
| chr11_+_62496114 | 0.18 |
ENST00000532583.1
|
TTC9C
|
tetratricopeptide repeat domain 9C |
| chr5_+_148786518 | 0.18 |
ENST00000518014.1
ENST00000505340.1 ENST00000509909.1 |
AC131025.8
MIR143HG
|
AC131025.8 MIR143 host gene (non-protein coding) |
| chr1_-_27286897 | 0.17 |
ENST00000320567.5
|
C1orf172
|
chromosome 1 open reading frame 172 |
| chr3_+_19988566 | 0.17 |
ENST00000273047.4
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr11_-_6462210 | 0.17 |
ENST00000265983.3
|
HPX
|
hemopexin |
| chr11_+_62495997 | 0.17 |
ENST00000316461.4
|
TTC9C
|
tetratricopeptide repeat domain 9C |
| chr19_-_45926739 | 0.17 |
ENST00000589381.1
ENST00000591636.1 ENST00000013807.5 ENST00000592023.1 |
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr6_-_39902160 | 0.16 |
ENST00000340692.5
|
MOCS1
|
molybdenum cofactor synthesis 1 |
| chr12_+_56415100 | 0.16 |
ENST00000547791.1
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr8_-_13134045 | 0.16 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr18_-_3845293 | 0.16 |
ENST00000400145.2
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr14_+_79746249 | 0.16 |
ENST00000428277.2
|
NRXN3
|
neurexin 3 |
| chrX_-_44402231 | 0.15 |
ENST00000378045.4
|
FUNDC1
|
FUN14 domain containing 1 |
| chr2_-_114514181 | 0.15 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chr1_-_22109484 | 0.15 |
ENST00000529637.1
|
USP48
|
ubiquitin specific peptidase 48 |
| chr13_-_36944307 | 0.15 |
ENST00000355182.4
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr15_-_90222642 | 0.14 |
ENST00000430628.2
|
PLIN1
|
perilipin 1 |
| chr5_+_132009675 | 0.14 |
ENST00000231449.2
ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr15_-_90222610 | 0.14 |
ENST00000300055.5
|
PLIN1
|
perilipin 1 |
| chr12_-_54652060 | 0.14 |
ENST00000552562.1
|
CBX5
|
chromobox homolog 5 |
| chr18_-_19748331 | 0.14 |
ENST00000584201.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr16_-_65155979 | 0.14 |
ENST00000562325.1
ENST00000268603.4 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr12_-_120884175 | 0.14 |
ENST00000546954.1
|
TRIAP1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr10_+_103113840 | 0.13 |
ENST00000393441.4
ENST00000408038.2 |
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr17_-_47287928 | 0.13 |
ENST00000507680.1
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr6_-_100016678 | 0.13 |
ENST00000523799.1
ENST00000520429.1 |
CCNC
|
cyclin C |
| chr3_+_19988885 | 0.13 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr1_+_101361782 | 0.13 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr14_+_79745746 | 0.13 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr16_-_65155833 | 0.12 |
ENST00000566827.1
ENST00000394156.3 ENST00000562998.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr3_-_182698381 | 0.12 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr7_-_102232891 | 0.12 |
ENST00000514917.2
|
RP11-514P8.7
|
RP11-514P8.7 |
| chrX_+_153409678 | 0.12 |
ENST00000369951.4
|
OPN1LW
|
opsin 1 (cone pigments), long-wave-sensitive |
| chr1_-_156460391 | 0.12 |
ENST00000360595.3
|
MEF2D
|
myocyte enhancer factor 2D |
| chr8_+_79428539 | 0.12 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr2_-_113999260 | 0.11 |
ENST00000468980.2
|
PAX8
|
paired box 8 |
| chr2_-_87248975 | 0.11 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr1_-_11863571 | 0.11 |
ENST00000376583.3
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr16_+_19567016 | 0.11 |
ENST00000251143.5
ENST00000417362.2 ENST00000567245.1 ENST00000513947.4 |
C16orf62
|
chromosome 16 open reading frame 62 |
| chr17_+_18761417 | 0.11 |
ENST00000419284.2
ENST00000268835.2 ENST00000412418.1 ENST00000575228.1 ENST00000575102.1 ENST00000536323.1 |
PRPSAP2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr4_+_84377198 | 0.10 |
ENST00000507349.1
ENST00000509970.1 ENST00000505719.1 |
MRPS18C
|
mitochondrial ribosomal protein S18C |
| chr1_+_202091980 | 0.10 |
ENST00000367282.5
|
GPR37L1
|
G protein-coupled receptor 37 like 1 |
| chr2_+_189839046 | 0.09 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr6_-_100016527 | 0.09 |
ENST00000523985.1
ENST00000518714.1 ENST00000520371.1 |
CCNC
|
cyclin C |
| chr2_-_26700900 | 0.09 |
ENST00000338581.6
ENST00000339598.3 ENST00000402415.3 |
OTOF
|
otoferlin |
| chr3_-_69129501 | 0.09 |
ENST00000540295.1
ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3
|
ubiquitin-like modifier activating enzyme 3 |
| chr17_-_79817091 | 0.09 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr12_-_57030115 | 0.09 |
ENST00000379441.3
ENST00000179765.5 ENST00000551812.1 |
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr4_-_84376953 | 0.09 |
ENST00000510985.1
ENST00000295488.3 |
HELQ
|
helicase, POLQ-like |
| chr14_-_44976474 | 0.08 |
ENST00000340446.4
|
FSCB
|
fibrous sheath CABYR binding protein |
| chr16_+_9185450 | 0.08 |
ENST00000327827.7
|
C16orf72
|
chromosome 16 open reading frame 72 |
| chr16_-_72206034 | 0.08 |
ENST00000537465.1
ENST00000237353.10 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr1_-_67896069 | 0.08 |
ENST00000370995.2
ENST00000361219.6 |
SERBP1
|
SERPINE1 mRNA binding protein 1 |
| chr2_-_128642434 | 0.07 |
ENST00000393001.1
|
AMMECR1L
|
AMMECR1-like |
| chr17_-_46691990 | 0.07 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr17_+_33474860 | 0.07 |
ENST00000394570.2
|
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr17_+_33474826 | 0.06 |
ENST00000268876.5
ENST00000433649.1 ENST00000378449.1 |
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr3_-_183145765 | 0.06 |
ENST00000473233.1
|
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.5 | 1.6 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.4 | 2.3 | GO:2000391 | positive regulation of neutrophil extravasation(GO:2000391) |
| 0.3 | 3.0 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.3 | 1.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.3 | 1.5 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 0.7 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.2 | 1.0 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.2 | 0.7 | GO:0014806 | negative regulation of muscle hyperplasia(GO:0014740) smooth muscle hyperplasia(GO:0014806) |
| 0.2 | 0.6 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 1.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 0.6 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 1.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 1.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.6 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 5.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 2.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.4 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.6 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.1 | 0.7 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 0.7 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 0.6 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.7 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 | 1.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.3 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.8 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.4 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 1.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 1.0 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.1 | 0.2 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.1 | 0.3 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.7 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.4 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.7 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.8 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.3 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.6 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.3 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.1 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.2 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.9 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 1.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 1.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 1.0 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.2 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 1.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:2001138 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.0 | 0.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.5 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.4 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.0 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 0.6 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 3.0 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.3 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 1.8 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.7 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 1.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.2 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 1.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.3 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.3 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 1.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 2.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.7 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 2.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 8.2 | GO:0015629 | actin cytoskeleton(GO:0015629) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.6 | 1.8 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.3 | 1.6 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.3 | 1.5 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 1.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 0.7 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.2 | 0.6 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.2 | 0.7 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 1.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.6 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 2.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 2.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 1.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.5 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.7 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 1.0 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 1.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 1.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 1.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 6.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 2.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.7 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 3.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 5.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 3.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 1.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 1.2 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 1.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |