Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
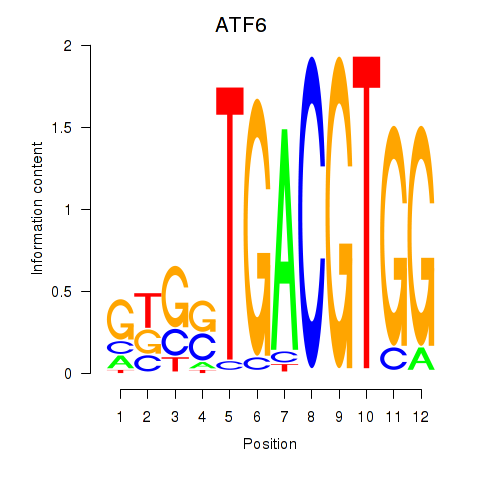
Results for ATF6
Z-value: 0.62
Transcription factors associated with ATF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF6
|
ENSG00000118217.5 | activating transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF6 | hg19_v2_chr1_+_161736072_161736093 | -0.21 | 2.6e-01 | Click! |
Activity profile of ATF6 motif
Sorted Z-values of ATF6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_100914781 | 1.08 |
ENST00000431597.1
ENST00000458024.1 ENST00000413506.1 ENST00000440675.1 ENST00000328766.5 ENST00000356824.4 |
ARMCX2
|
armadillo repeat containing, X-linked 2 |
| chr19_+_35630926 | 0.91 |
ENST00000588081.1
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr19_+_35630628 | 0.90 |
ENST00000588715.1
ENST00000588607.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr10_-_118032979 | 0.62 |
ENST00000355422.6
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr7_-_19157248 | 0.55 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1 |
| chr12_+_56661033 | 0.51 |
ENST00000433805.2
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr12_+_56660633 | 0.48 |
ENST00000308197.5
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr15_+_72947079 | 0.40 |
ENST00000421285.3
|
GOLGA6B
|
golgin A6 family, member B |
| chrX_-_101397433 | 0.39 |
ENST00000372774.3
|
TCEAL6
|
transcription elongation factor A (SII)-like 6 |
| chrY_+_2803322 | 0.39 |
ENST00000383052.1
ENST00000155093.3 ENST00000449237.1 ENST00000443793.1 |
ZFY
|
zinc finger protein, Y-linked |
| chr3_+_100211412 | 0.38 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr14_+_65016620 | 0.37 |
ENST00000298705.1
|
PPP1R36
|
protein phosphatase 1, regulatory subunit 36 |
| chr2_-_74692473 | 0.37 |
ENST00000535045.1
ENST00000409065.1 ENST00000414701.1 ENST00000448666.1 ENST00000233616.4 ENST00000452063.2 |
MOGS
|
mannosyl-oligosaccharide glucosidase |
| chr3_+_184053703 | 0.37 |
ENST00000450976.1
ENST00000418281.1 ENST00000340957.5 ENST00000433578.1 |
FAM131A
|
family with sequence similarity 131, member A |
| chr14_+_68086515 | 0.35 |
ENST00000261783.3
|
ARG2
|
arginase 2 |
| chr7_+_102073966 | 0.35 |
ENST00000495936.1
ENST00000356387.2 ENST00000478730.2 ENST00000468241.1 ENST00000403646.3 |
ORAI2
|
ORAI calcium release-activated calcium modulator 2 |
| chr14_-_106092403 | 0.34 |
ENST00000390543.2
|
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker) |
| chr9_+_100000717 | 0.34 |
ENST00000375205.2
ENST00000357054.1 ENST00000395220.1 ENST00000375202.2 ENST00000411667.2 |
CCDC180
|
coiled-coil domain containing 180 |
| chr3_+_129158926 | 0.32 |
ENST00000347300.2
ENST00000296266.3 |
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr3_+_129159039 | 0.32 |
ENST00000507564.1
ENST00000431818.2 ENST00000504021.1 ENST00000349441.2 ENST00000348417.2 ENST00000440957.2 |
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr6_+_159291090 | 0.31 |
ENST00000367073.4
ENST00000608817.1 |
C6orf99
|
chromosome 6 open reading frame 99 |
| chr6_+_159290917 | 0.30 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr3_+_184056614 | 0.30 |
ENST00000453072.1
|
FAM131A
|
family with sequence similarity 131, member A |
| chr9_-_79520989 | 0.29 |
ENST00000376713.3
ENST00000376718.3 ENST00000428286.1 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr14_+_105190514 | 0.28 |
ENST00000330877.2
|
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr1_-_94050668 | 0.27 |
ENST00000539242.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr20_+_35090150 | 0.27 |
ENST00000340491.4
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4 |
| chr12_+_117176113 | 0.27 |
ENST00000319176.7
|
RNFT2
|
ring finger protein, transmembrane 2 |
| chr2_-_216946500 | 0.27 |
ENST00000265322.7
|
PECR
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr5_+_94890778 | 0.26 |
ENST00000380009.4
|
ARSK
|
arylsulfatase family, member K |
| chr2_-_47572105 | 0.25 |
ENST00000419035.1
ENST00000448713.1 ENST00000450550.1 ENST00000413185.2 |
AC073283.4
|
AC073283.4 |
| chr1_-_151254362 | 0.25 |
ENST00000447795.2
|
RP11-126K1.2
|
Uncharacterized protein |
| chr18_+_56530794 | 0.25 |
ENST00000590285.1
ENST00000586085.1 ENST00000589288.1 |
ZNF532
|
zinc finger protein 532 |
| chrX_+_102862834 | 0.25 |
ENST00000372627.5
ENST00000243286.3 |
TCEAL3
|
transcription elongation factor A (SII)-like 3 |
| chr22_+_38864041 | 0.25 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr19_-_42927251 | 0.24 |
ENST00000597001.1
|
LIPE
|
lipase, hormone-sensitive |
| chr15_-_74374891 | 0.24 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family, member A |
| chr1_-_15850676 | 0.23 |
ENST00000440484.1
ENST00000333868.5 |
CASP9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr5_+_78532003 | 0.22 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr11_-_119234876 | 0.22 |
ENST00000525735.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr6_+_24775641 | 0.22 |
ENST00000378054.2
ENST00000476555.1 |
GMNN
|
geminin, DNA replication inhibitor |
| chr8_+_22422749 | 0.22 |
ENST00000523900.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr6_+_24775153 | 0.22 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr10_+_35415851 | 0.22 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chr21_-_38445011 | 0.22 |
ENST00000464265.1
ENST00000399102.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr22_+_42394780 | 0.21 |
ENST00000328823.9
|
WBP2NL
|
WBP2 N-terminal like |
| chr19_-_40724246 | 0.21 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B |
| chr1_-_185126037 | 0.21 |
ENST00000367506.5
ENST00000367504.3 |
TRMT1L
|
tRNA methyltransferase 1 homolog (S. cerevisiae)-like |
| chr17_+_33914460 | 0.21 |
ENST00000537622.2
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr11_+_61717842 | 0.20 |
ENST00000449131.2
|
BEST1
|
bestrophin 1 |
| chr4_+_128982416 | 0.20 |
ENST00000326639.6
|
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr11_-_33183048 | 0.20 |
ENST00000438862.2
|
CSTF3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3, 77kDa |
| chr3_+_33318914 | 0.20 |
ENST00000484457.1
ENST00000538892.1 ENST00000538181.1 ENST00000446237.3 ENST00000507198.1 |
FBXL2
|
F-box and leucine-rich repeat protein 2 |
| chr17_+_33914276 | 0.20 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr8_-_38126675 | 0.20 |
ENST00000531823.1
ENST00000534339.1 ENST00000524616.1 ENST00000422581.2 ENST00000424479.2 ENST00000419686.2 |
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr17_+_6554971 | 0.19 |
ENST00000391428.2
|
C17orf100
|
chromosome 17 open reading frame 100 |
| chr17_-_57184260 | 0.19 |
ENST00000376149.3
ENST00000393066.3 |
TRIM37
|
tripartite motif containing 37 |
| chr9_-_89562104 | 0.19 |
ENST00000298743.7
|
GAS1
|
growth arrest-specific 1 |
| chr8_-_124408652 | 0.18 |
ENST00000287394.5
|
ATAD2
|
ATPase family, AAA domain containing 2 |
| chr5_+_63802109 | 0.18 |
ENST00000334025.2
|
RGS7BP
|
regulator of G-protein signaling 7 binding protein |
| chr7_+_30811004 | 0.18 |
ENST00000265299.6
|
FAM188B
|
family with sequence similarity 188, member B |
| chrX_+_122318006 | 0.18 |
ENST00000371266.1
ENST00000264357.5 |
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr11_+_62496114 | 0.18 |
ENST00000532583.1
|
TTC9C
|
tetratricopeptide repeat domain 9C |
| chr7_-_98467629 | 0.17 |
ENST00000339375.4
|
TMEM130
|
transmembrane protein 130 |
| chr14_-_106068065 | 0.17 |
ENST00000390541.2
|
IGHE
|
immunoglobulin heavy constant epsilon |
| chr15_+_75575176 | 0.17 |
ENST00000434739.3
|
GOLGA6D
|
golgin A6 family, member D |
| chr17_-_66951474 | 0.17 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr18_-_46987000 | 0.17 |
ENST00000442713.2
ENST00000269445.6 |
DYM
|
dymeclin |
| chr10_+_35416090 | 0.17 |
ENST00000354759.3
|
CREM
|
cAMP responsive element modulator |
| chr14_+_76776957 | 0.17 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr2_+_219745020 | 0.17 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr1_+_221051699 | 0.16 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr9_+_100818976 | 0.16 |
ENST00000210444.5
|
NANS
|
N-acetylneuraminic acid synthase |
| chr7_-_98467489 | 0.16 |
ENST00000416379.2
|
TMEM130
|
transmembrane protein 130 |
| chr21_-_38445470 | 0.16 |
ENST00000399098.1
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr4_-_24914576 | 0.16 |
ENST00000502801.1
ENST00000428116.2 |
CCDC149
|
coiled-coil domain containing 149 |
| chr7_-_98467543 | 0.16 |
ENST00000345589.4
|
TMEM130
|
transmembrane protein 130 |
| chr4_+_85504075 | 0.16 |
ENST00000295887.5
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr4_-_119757239 | 0.16 |
ENST00000280551.6
|
SEC24D
|
SEC24 family member D |
| chr17_-_49337392 | 0.16 |
ENST00000376381.2
ENST00000586178.1 |
MBTD1
|
mbt domain containing 1 |
| chr1_-_113498616 | 0.16 |
ENST00000433570.4
ENST00000538576.1 ENST00000458229.1 |
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr5_+_126112794 | 0.16 |
ENST00000261366.5
ENST00000395354.1 |
LMNB1
|
lamin B1 |
| chr8_+_142138711 | 0.16 |
ENST00000518347.1
ENST00000262585.2 ENST00000424248.1 ENST00000519811.1 ENST00000520986.1 ENST00000523058.1 |
DENND3
|
DENN/MADD domain containing 3 |
| chr11_-_113746277 | 0.16 |
ENST00000003302.4
ENST00000545540.1 |
USP28
|
ubiquitin specific peptidase 28 |
| chr10_+_35415719 | 0.15 |
ENST00000474362.1
ENST00000374721.3 |
CREM
|
cAMP responsive element modulator |
| chr9_+_114393581 | 0.15 |
ENST00000313525.3
|
DNAJC25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr7_+_5632436 | 0.15 |
ENST00000340250.6
ENST00000382361.3 |
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
| chr20_+_32951070 | 0.15 |
ENST00000535650.1
ENST00000262650.6 |
ITCH
|
itchy E3 ubiquitin protein ligase |
| chr17_+_42977122 | 0.15 |
ENST00000412523.2
ENST00000331733.4 ENST00000417826.2 |
FAM187A
CCDC103
|
family with sequence similarity 187, member A coiled-coil domain containing 103 |
| chr7_-_8302164 | 0.15 |
ENST00000447326.1
ENST00000406470.2 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr2_-_211036051 | 0.15 |
ENST00000418791.1
ENST00000452086.1 ENST00000281772.9 |
KANSL1L
|
KAT8 regulatory NSL complex subunit 1-like |
| chr6_-_135818844 | 0.15 |
ENST00000524469.1
ENST00000367800.4 ENST00000327035.6 ENST00000457866.2 ENST00000265602.6 |
AHI1
|
Abelson helper integration site 1 |
| chr1_-_113498943 | 0.14 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr2_-_60780702 | 0.14 |
ENST00000359629.5
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr7_-_8301682 | 0.14 |
ENST00000396675.3
ENST00000430867.1 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr10_-_105218645 | 0.14 |
ENST00000329905.5
|
CALHM1
|
calcium homeostasis modulator 1 |
| chr2_-_242212227 | 0.14 |
ENST00000427007.1
ENST00000458564.1 ENST00000452065.1 ENST00000427183.2 ENST00000426343.1 ENST00000422080.1 ENST00000449504.1 ENST00000449864.1 ENST00000391975.1 |
HDLBP
|
high density lipoprotein binding protein |
| chr5_+_34656569 | 0.14 |
ENST00000428746.2
|
RAI14
|
retinoic acid induced 14 |
| chr1_+_185126291 | 0.14 |
ENST00000367500.4
|
SWT1
|
SWT1 RNA endoribonuclease homolog (S. cerevisiae) |
| chr8_-_38126635 | 0.14 |
ENST00000529359.1
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr19_-_46916805 | 0.14 |
ENST00000307522.3
|
CCDC8
|
coiled-coil domain containing 8 |
| chrX_+_122318113 | 0.14 |
ENST00000371264.3
|
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr20_+_34042962 | 0.14 |
ENST00000446710.1
ENST00000420564.1 |
CEP250
|
centrosomal protein 250kDa |
| chr9_-_77643189 | 0.14 |
ENST00000376837.3
|
C9orf41
|
chromosome 9 open reading frame 41 |
| chr21_-_38445297 | 0.14 |
ENST00000430792.1
ENST00000399103.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr7_-_86849883 | 0.14 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr1_+_110527308 | 0.14 |
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr10_-_27444143 | 0.14 |
ENST00000477432.1
|
YME1L1
|
YME1-like 1 ATPase |
| chr16_+_19125252 | 0.13 |
ENST00000566735.1
ENST00000381440.3 |
ITPRIPL2
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 2 |
| chr9_+_131038425 | 0.13 |
ENST00000320188.5
ENST00000608796.1 ENST00000419867.2 ENST00000418976.1 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr7_-_8301768 | 0.13 |
ENST00000265577.7
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr13_-_36920615 | 0.13 |
ENST00000494062.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr7_-_8301869 | 0.13 |
ENST00000402384.3
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr2_-_27712583 | 0.13 |
ENST00000260570.3
ENST00000359466.6 ENST00000416524.2 |
IFT172
|
intraflagellar transport 172 homolog (Chlamydomonas) |
| chr5_+_34656331 | 0.13 |
ENST00000265109.3
|
RAI14
|
retinoic acid induced 14 |
| chr5_+_56469843 | 0.13 |
ENST00000514387.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr12_+_7282795 | 0.13 |
ENST00000266546.6
|
CLSTN3
|
calsyntenin 3 |
| chr11_-_113746212 | 0.13 |
ENST00000537642.1
ENST00000537706.1 ENST00000544750.1 ENST00000260188.5 ENST00000540925.1 |
USP28
|
ubiquitin specific peptidase 28 |
| chr19_+_16186903 | 0.13 |
ENST00000588507.1
|
TPM4
|
tropomyosin 4 |
| chr10_+_35415978 | 0.13 |
ENST00000429130.3
ENST00000469949.2 ENST00000460270.1 |
CREM
|
cAMP responsive element modulator |
| chr11_-_57417405 | 0.13 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr6_+_116692102 | 0.13 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase |
| chr4_+_668348 | 0.12 |
ENST00000511290.1
|
MYL5
|
myosin, light chain 5, regulatory |
| chr21_-_38445443 | 0.12 |
ENST00000360525.4
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr8_-_144623595 | 0.12 |
ENST00000262577.5
|
ZC3H3
|
zinc finger CCCH-type containing 3 |
| chr14_+_55518349 | 0.12 |
ENST00000395468.4
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr4_-_39640700 | 0.12 |
ENST00000295958.5
|
SMIM14
|
small integral membrane protein 14 |
| chr17_-_39968406 | 0.12 |
ENST00000393928.1
|
LEPREL4
|
leprecan-like 4 |
| chr13_+_53226963 | 0.12 |
ENST00000343788.6
ENST00000535397.1 ENST00000310528.8 |
SUGT1
|
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| chr7_-_100808843 | 0.12 |
ENST00000249330.2
|
VGF
|
VGF nerve growth factor inducible |
| chr15_-_82641706 | 0.12 |
ENST00000439287.4
|
GOLGA6L10
|
golgin A6 family-like 10 |
| chr15_-_52030293 | 0.12 |
ENST00000560491.1
ENST00000267838.3 |
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr6_-_33714752 | 0.12 |
ENST00000451316.1
|
IP6K3
|
inositol hexakisphosphate kinase 3 |
| chr12_-_76953453 | 0.12 |
ENST00000549570.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr20_-_22559211 | 0.11 |
ENST00000564492.1
|
LINC00261
|
long intergenic non-protein coding RNA 261 |
| chr8_+_61429728 | 0.11 |
ENST00000529579.1
|
RAB2A
|
RAB2A, member RAS oncogene family |
| chr13_-_36920872 | 0.11 |
ENST00000451493.1
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr7_-_100808394 | 0.11 |
ENST00000445482.2
|
VGF
|
VGF nerve growth factor inducible |
| chr7_+_65540853 | 0.11 |
ENST00000380839.4
ENST00000395332.3 ENST00000362000.5 ENST00000395331.3 |
ASL
|
argininosuccinate lyase |
| chr5_+_56469775 | 0.11 |
ENST00000424459.3
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr2_+_216176761 | 0.11 |
ENST00000540518.1
ENST00000435675.1 |
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr19_-_39523165 | 0.11 |
ENST00000509137.2
ENST00000292853.4 |
FBXO27
|
F-box protein 27 |
| chr1_-_51425772 | 0.11 |
ENST00000371778.4
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr15_-_72668185 | 0.11 |
ENST00000457859.2
ENST00000566304.1 ENST00000567159.1 ENST00000429918.2 |
HEXA
|
hexosaminidase A (alpha polypeptide) |
| chr7_+_16793160 | 0.11 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr2_+_28615669 | 0.11 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr22_-_39096661 | 0.11 |
ENST00000216039.5
|
JOSD1
|
Josephin domain containing 1 |
| chr4_+_1873155 | 0.11 |
ENST00000507820.1
ENST00000514045.1 |
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr1_-_25573937 | 0.11 |
ENST00000417642.2
ENST00000431849.2 |
C1orf63
|
chromosome 1 open reading frame 63 |
| chr1_-_15850839 | 0.10 |
ENST00000348549.5
ENST00000546424.1 |
CASP9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr2_+_216176540 | 0.10 |
ENST00000236959.9
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr4_+_128982490 | 0.10 |
ENST00000394288.3
ENST00000432347.2 ENST00000264584.5 ENST00000441387.1 ENST00000427266.1 ENST00000354456.3 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr5_-_132299290 | 0.10 |
ENST00000378595.3
|
AFF4
|
AF4/FMR2 family, member 4 |
| chr7_+_65540780 | 0.10 |
ENST00000304874.9
|
ASL
|
argininosuccinate lyase |
| chr2_+_183580954 | 0.10 |
ENST00000264065.7
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr3_+_50284321 | 0.10 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr4_-_39640513 | 0.10 |
ENST00000511809.1
ENST00000505729.1 |
SMIM14
|
small integral membrane protein 14 |
| chr6_+_33168637 | 0.10 |
ENST00000374677.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr12_+_1100449 | 0.10 |
ENST00000360905.4
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr6_+_132873832 | 0.10 |
ENST00000275200.1
|
TAAR8
|
trace amine associated receptor 8 |
| chrX_-_100662881 | 0.10 |
ENST00000218516.3
|
GLA
|
galactosidase, alpha |
| chr6_+_127587755 | 0.10 |
ENST00000368314.1
ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146
|
ring finger protein 146 |
| chr6_+_26204825 | 0.10 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr20_-_44420507 | 0.10 |
ENST00000243938.4
|
WFDC3
|
WAP four-disulfide core domain 3 |
| chr12_+_49372251 | 0.10 |
ENST00000293549.3
|
WNT1
|
wingless-type MMTV integration site family, member 1 |
| chr2_-_190649082 | 0.10 |
ENST00000392350.3
ENST00000392349.4 |
ORMDL1
|
ORM1-like 1 (S. cerevisiae) |
| chr11_+_28131821 | 0.10 |
ENST00000379199.2
ENST00000303459.6 |
METTL15
|
methyltransferase like 15 |
| chr12_-_56709674 | 0.10 |
ENST00000551286.1
ENST00000549318.1 |
CNPY2
RP11-977G19.10
|
canopy FGF signaling regulator 2 Uncharacterized protein |
| chr1_+_156830607 | 0.10 |
ENST00000368196.3
|
NTRK1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr16_+_22217577 | 0.10 |
ENST00000263026.5
|
EEF2K
|
eukaryotic elongation factor-2 kinase |
| chr19_-_18391708 | 0.10 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr17_+_65821636 | 0.10 |
ENST00000544778.2
|
BPTF
|
bromodomain PHD finger transcription factor |
| chr12_+_117176090 | 0.10 |
ENST00000257575.4
ENST00000407967.3 ENST00000392549.2 |
RNFT2
|
ring finger protein, transmembrane 2 |
| chr3_-_156877997 | 0.09 |
ENST00000295926.3
|
CCNL1
|
cyclin L1 |
| chr6_+_31865552 | 0.09 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chr6_+_33168597 | 0.09 |
ENST00000374675.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr20_+_32951041 | 0.09 |
ENST00000374864.4
|
ITCH
|
itchy E3 ubiquitin protein ligase |
| chr3_-_195270162 | 0.09 |
ENST00000438848.1
ENST00000328432.3 |
PPP1R2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr1_-_25573977 | 0.09 |
ENST00000243189.7
|
C1orf63
|
chromosome 1 open reading frame 63 |
| chr17_-_39968855 | 0.09 |
ENST00000355468.3
ENST00000590496.1 |
LEPREL4
|
leprecan-like 4 |
| chr10_-_74856608 | 0.09 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr14_-_81687575 | 0.09 |
ENST00000434192.2
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr13_-_36920420 | 0.09 |
ENST00000438666.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr9_-_88356733 | 0.09 |
ENST00000376083.3
|
AGTPBP1
|
ATP/GTP binding protein 1 |
| chr8_-_75233563 | 0.09 |
ENST00000342232.4
|
JPH1
|
junctophilin 1 |
| chr17_+_17876127 | 0.09 |
ENST00000582416.1
ENST00000313838.8 ENST00000411504.2 ENST00000581264.1 ENST00000399187.1 ENST00000479684.2 ENST00000584166.1 ENST00000585108.1 ENST00000399182.1 ENST00000579977.1 |
LRRC48
|
leucine rich repeat containing 48 |
| chr16_+_19729586 | 0.09 |
ENST00000564186.1
ENST00000541926.1 ENST00000433597.2 |
IQCK
|
IQ motif containing K |
| chr10_+_102759045 | 0.09 |
ENST00000370220.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr1_-_165325939 | 0.09 |
ENST00000342310.3
|
LMX1A
|
LIM homeobox transcription factor 1, alpha |
| chr1_+_156830678 | 0.09 |
ENST00000524377.1
ENST00000358660.3 |
NTRK1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr14_-_50087312 | 0.09 |
ENST00000298289.6
|
RPL36AL
|
ribosomal protein L36a-like |
| chr12_-_49412525 | 0.09 |
ENST00000551121.1
ENST00000552212.1 ENST00000548605.1 ENST00000548950.1 ENST00000547125.1 |
PRKAG1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr1_+_114472481 | 0.09 |
ENST00000369555.2
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr4_+_128554081 | 0.09 |
ENST00000335251.6
ENST00000296461.5 |
INTU
|
inturned planar cell polarity protein |
| chr12_+_1100370 | 0.09 |
ENST00000543086.3
ENST00000546231.2 ENST00000397203.2 |
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr4_-_54930790 | 0.08 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr2_-_201753980 | 0.08 |
ENST00000443398.1
ENST00000286175.8 ENST00000409449.1 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr3_-_129158850 | 0.08 |
ENST00000503197.1
ENST00000249910.1 ENST00000429544.2 ENST00000507208.1 |
MBD4
|
methyl-CpG binding domain protein 4 |
| chr11_+_47430133 | 0.08 |
ENST00000531974.1
ENST00000531419.1 ENST00000531865.1 ENST00000362021.4 ENST00000354884.4 |
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr11_-_33183006 | 0.08 |
ENST00000524827.1
ENST00000323959.4 ENST00000431742.2 |
CSTF3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3, 77kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.2 | 0.6 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.2 | 0.6 | GO:0035720 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.3 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.1 | 0.3 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.4 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.4 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.2 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.1 | 0.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.2 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.2 | GO:0035519 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) pronephric nephron development(GO:0039019) |
| 0.0 | 0.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:2000173 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.6 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 1.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0061183 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.1 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.0 | 0.3 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.6 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.1 | GO:1902824 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.0 | 0.1 | GO:1990144 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.0 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:1903984 | negative regulation of ribosome biogenesis(GO:0090071) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0061525 | hindgut development(GO:0061525) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.4 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 | 0.0 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) positive regulation of NK T cell differentiation(GO:0051138) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 1.3 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.0 | GO:0035993 | subthalamic nucleus development(GO:0021763) deltoid tuberosity development(GO:0035993) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 1.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.6 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.2 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.2 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0030849 | autosome(GO:0030849) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.4 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.3 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.1 | 0.3 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.3 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.6 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.2 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.1 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 1.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.4 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.0 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.0 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.0 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.0 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |