Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
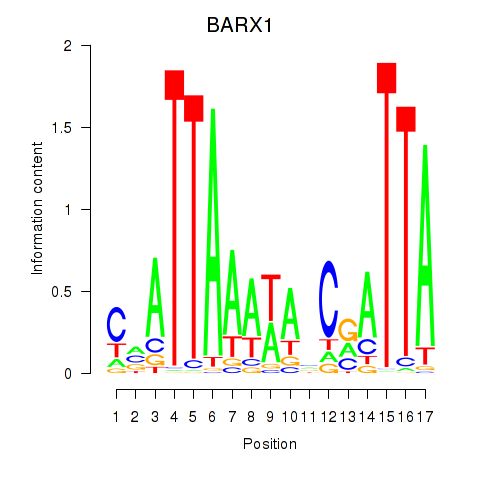
Results for BARX1
Z-value: 0.85
Transcription factors associated with BARX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BARX1
|
ENSG00000131668.9 | BARX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BARX1 | hg19_v2_chr9_-_96717654_96717666 | -0.23 | 2.1e-01 | Click! |
Activity profile of BARX1 motif
Sorted Z-values of BARX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_56757329 | 3.38 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr7_-_16921601 | 3.06 |
ENST00000402239.3
ENST00000310398.2 ENST00000414935.1 |
AGR3
|
anterior gradient 3 |
| chr6_+_32407619 | 2.47 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr11_+_61976137 | 2.32 |
ENST00000244930.4
|
SCGB2A1
|
secretoglobin, family 2A, member 1 |
| chr12_+_9980113 | 2.06 |
ENST00000537723.1
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr4_+_52917451 | 1.78 |
ENST00000295213.4
ENST00000419395.2 |
SPATA18
|
spermatogenesis associated 18 |
| chr12_+_20963632 | 1.59 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr6_+_163148161 | 1.59 |
ENST00000337019.3
ENST00000366889.2 |
PACRG
|
PARK2 co-regulated |
| chr12_-_71551652 | 1.59 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr5_+_121465207 | 1.54 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr6_+_131958436 | 1.43 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr15_+_71228826 | 1.40 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr12_+_20963647 | 1.39 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr2_+_228736335 | 1.37 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr12_-_71551868 | 1.36 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr21_-_35884573 | 1.21 |
ENST00000399286.2
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1 |
| chr4_-_70518941 | 1.21 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr21_-_43735628 | 1.12 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr5_-_135290705 | 1.09 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr17_+_44588877 | 1.08 |
ENST00000576629.1
|
LRRC37A2
|
leucine rich repeat containing 37, member A2 |
| chr2_+_109403193 | 1.08 |
ENST00000412964.2
ENST00000295124.4 |
CCDC138
|
coiled-coil domain containing 138 |
| chr12_+_9980069 | 1.07 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr3_-_197300194 | 1.04 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr7_+_138818490 | 1.02 |
ENST00000430935.1
ENST00000495038.1 ENST00000474035.2 ENST00000478836.2 ENST00000464848.1 ENST00000343187.4 |
TTC26
|
tetratricopeptide repeat domain 26 |
| chr10_+_51549498 | 0.97 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr11_+_94245617 | 0.92 |
ENST00000542198.1
|
RP11-867G2.2
|
long intergenic non-protein coding RNA 1171 |
| chr22_-_50970919 | 0.92 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr5_+_140213815 | 0.91 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr1_+_36549676 | 0.91 |
ENST00000207457.3
|
TEKT2
|
tektin 2 (testicular) |
| chr5_+_156696362 | 0.90 |
ENST00000377576.3
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr1_-_152131703 | 0.88 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr1_-_150738261 | 0.88 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr3_+_97483572 | 0.88 |
ENST00000335979.2
ENST00000394206.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr6_-_150212029 | 0.87 |
ENST00000529948.1
ENST00000357183.4 ENST00000367363.3 |
RAET1E
|
retinoic acid early transcript 1E |
| chr7_+_13141097 | 0.85 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr11_+_60467047 | 0.83 |
ENST00000300226.2
|
MS4A8
|
membrane-spanning 4-domains, subfamily A, member 8 |
| chr12_+_45609797 | 0.82 |
ENST00000425752.2
|
ANO6
|
anoctamin 6 |
| chr17_-_19266045 | 0.81 |
ENST00000395616.3
|
B9D1
|
B9 protein domain 1 |
| chr12_-_58329819 | 0.80 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr5_-_43412418 | 0.80 |
ENST00000537013.1
ENST00000361115.4 |
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr3_+_160559931 | 0.79 |
ENST00000464260.1
ENST00000295839.9 |
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr5_-_124081008 | 0.76 |
ENST00000306315.5
|
ZNF608
|
zinc finger protein 608 |
| chr4_+_41540160 | 0.76 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_-_9994021 | 0.75 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr16_+_23765948 | 0.75 |
ENST00000300113.2
|
CHP2
|
calcineurin-like EF-hand protein 2 |
| chr10_-_79398250 | 0.74 |
ENST00000286627.5
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr6_+_159071015 | 0.73 |
ENST00000360448.3
|
SYTL3
|
synaptotagmin-like 3 |
| chr9_+_139874683 | 0.72 |
ENST00000444903.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr1_+_61330931 | 0.70 |
ENST00000371191.1
|
NFIA
|
nuclear factor I/A |
| chr4_-_38806404 | 0.70 |
ENST00000308979.2
ENST00000505940.1 ENST00000515861.1 |
TLR1
|
toll-like receptor 1 |
| chr12_+_21207503 | 0.69 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr19_-_14992264 | 0.68 |
ENST00000327462.2
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17 |
| chr10_+_114135952 | 0.67 |
ENST00000356116.1
ENST00000433418.1 ENST00000354273.4 |
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr13_-_86373536 | 0.66 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr3_-_3151664 | 0.65 |
ENST00000256452.3
ENST00000311981.8 ENST00000430514.2 ENST00000456302.1 |
IL5RA
|
interleukin 5 receptor, alpha |
| chr10_-_79398127 | 0.65 |
ENST00000372443.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chrX_-_103499602 | 0.64 |
ENST00000372588.4
|
ESX1
|
ESX homeobox 1 |
| chr5_+_140235469 | 0.64 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr10_-_79397740 | 0.63 |
ENST00000372440.1
ENST00000480683.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr17_-_19265982 | 0.63 |
ENST00000268841.6
ENST00000261499.4 ENST00000575478.1 |
B9D1
|
B9 protein domain 1 |
| chr12_-_49582978 | 0.63 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr8_+_94767072 | 0.63 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr6_-_49712123 | 0.62 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr20_+_58571419 | 0.62 |
ENST00000244049.3
ENST00000350849.6 ENST00000456106.1 |
CDH26
|
cadherin 26 |
| chr10_+_90660832 | 0.62 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr1_-_19615744 | 0.59 |
ENST00000361640.4
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr10_+_90354503 | 0.59 |
ENST00000531458.1
|
LIPJ
|
lipase, family member J |
| chr1_+_170904612 | 0.58 |
ENST00000367759.4
ENST00000367758.3 |
MROH9
|
maestro heat-like repeat family member 9 |
| chr13_+_24144796 | 0.57 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr1_-_95391315 | 0.57 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chrX_+_36246735 | 0.57 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr2_+_169312350 | 0.57 |
ENST00000305747.6
|
CERS6
|
ceramide synthase 6 |
| chr12_-_50290839 | 0.56 |
ENST00000552863.1
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr7_-_34978980 | 0.56 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr1_+_40840320 | 0.55 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr16_+_76587314 | 0.55 |
ENST00000563764.1
|
RP11-58C22.1
|
Uncharacterized protein |
| chr12_-_57328187 | 0.53 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr16_-_67517716 | 0.53 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chrX_-_13835147 | 0.52 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr16_+_89724188 | 0.52 |
ENST00000301031.4
ENST00000566204.1 ENST00000579310.1 |
SPATA33
|
spermatogenesis associated 33 |
| chr17_-_19265855 | 0.52 |
ENST00000440841.1
ENST00000395615.1 ENST00000461069.2 |
B9D1
|
B9 protein domain 1 |
| chr3_+_138340049 | 0.51 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr19_+_41281416 | 0.50 |
ENST00000597140.1
|
MIA
|
melanoma inhibitory activity |
| chr13_+_24144509 | 0.50 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr18_+_28956740 | 0.50 |
ENST00000308128.4
ENST00000359747.4 |
DSG4
|
desmoglein 4 |
| chr8_+_101170563 | 0.49 |
ENST00000520508.1
ENST00000388798.2 |
SPAG1
|
sperm associated antigen 1 |
| chr3_-_49314640 | 0.48 |
ENST00000436325.1
|
C3orf62
|
chromosome 3 open reading frame 62 |
| chr15_-_55657428 | 0.48 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr5_+_140220769 | 0.48 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chr5_+_147691979 | 0.47 |
ENST00000274565.4
|
SPINK7
|
serine peptidase inhibitor, Kazal type 7 (putative) |
| chr8_-_90996459 | 0.47 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr10_-_25305011 | 0.46 |
ENST00000331161.4
ENST00000376363.1 |
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr16_+_33204156 | 0.46 |
ENST00000398667.4
|
TP53TG3C
|
TP53 target 3C |
| chr1_-_118727781 | 0.46 |
ENST00000336338.5
|
SPAG17
|
sperm associated antigen 17 |
| chr15_+_75639296 | 0.46 |
ENST00000564500.1
ENST00000355059.4 ENST00000566752.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr1_-_89736434 | 0.46 |
ENST00000370459.3
|
GBP5
|
guanylate binding protein 5 |
| chr1_+_246887349 | 0.45 |
ENST00000366510.3
|
SCCPDH
|
saccharopine dehydrogenase (putative) |
| chr14_+_77582905 | 0.45 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr6_+_20534672 | 0.45 |
ENST00000274695.4
ENST00000378624.4 |
CDKAL1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr13_-_49975632 | 0.45 |
ENST00000457041.1
ENST00000355854.4 |
CAB39L
|
calcium binding protein 39-like |
| chr11_-_22647350 | 0.44 |
ENST00000327470.3
|
FANCF
|
Fanconi anemia, complementation group F |
| chr19_-_10491130 | 0.43 |
ENST00000530829.1
ENST00000529370.1 |
TYK2
|
tyrosine kinase 2 |
| chr17_+_26800296 | 0.42 |
ENST00000444914.3
ENST00000314669.5 |
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr19_-_9006766 | 0.42 |
ENST00000599436.1
|
MUC16
|
mucin 16, cell surface associated |
| chr15_+_72978521 | 0.42 |
ENST00000542334.1
ENST00000268057.4 |
BBS4
|
Bardet-Biedl syndrome 4 |
| chr2_-_175711133 | 0.42 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr6_-_52705641 | 0.42 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr4_+_26585686 | 0.41 |
ENST00000505206.1
ENST00000511789.1 |
TBC1D19
|
TBC1 domain family, member 19 |
| chr15_+_72978539 | 0.41 |
ENST00000539603.1
ENST00000569338.1 |
BBS4
|
Bardet-Biedl syndrome 4 |
| chr12_+_20968608 | 0.41 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr13_+_111855414 | 0.41 |
ENST00000375737.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr18_-_47721447 | 0.41 |
ENST00000285039.7
|
MYO5B
|
myosin VB |
| chr3_+_45927994 | 0.41 |
ENST00000357632.2
ENST00000395963.2 |
CCR9
|
chemokine (C-C motif) receptor 9 |
| chr2_+_191273052 | 0.40 |
ENST00000417958.1
ENST00000432036.1 ENST00000392328.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr16_-_28634874 | 0.40 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr3_-_146213722 | 0.40 |
ENST00000336685.2
ENST00000489015.1 |
PLSCR2
|
phospholipid scramblase 2 |
| chr3_-_93747425 | 0.40 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr12_+_41831485 | 0.39 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr3_+_25831567 | 0.39 |
ENST00000280701.3
ENST00000420173.2 |
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr17_-_34270697 | 0.39 |
ENST00000585556.1
|
LYZL6
|
lysozyme-like 6 |
| chr2_-_89310012 | 0.39 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr6_+_88106840 | 0.39 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164 |
| chr17_+_71228793 | 0.38 |
ENST00000426147.2
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr16_+_28648975 | 0.38 |
ENST00000529716.1
|
NPIPB8
|
nuclear pore complex interacting protein family, member B8 |
| chr5_-_55412774 | 0.38 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr15_-_52587945 | 0.38 |
ENST00000443683.2
ENST00000558479.1 ENST00000261839.7 |
MYO5C
|
myosin VC |
| chr18_+_61442629 | 0.38 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr9_-_116102562 | 0.38 |
ENST00000374193.4
ENST00000465979.1 |
WDR31
|
WD repeat domain 31 |
| chr5_+_140261703 | 0.38 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr3_+_171561127 | 0.37 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr4_+_69681710 | 0.37 |
ENST00000265403.7
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr10_+_116697946 | 0.37 |
ENST00000298746.3
|
TRUB1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr20_+_11898507 | 0.36 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr11_-_5255861 | 0.36 |
ENST00000380299.3
|
HBD
|
hemoglobin, delta |
| chr11_-_1629693 | 0.36 |
ENST00000399685.1
|
KRTAP5-3
|
keratin associated protein 5-3 |
| chr6_-_76072719 | 0.36 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr17_-_34270668 | 0.36 |
ENST00000293274.4
|
LYZL6
|
lysozyme-like 6 |
| chr5_+_140180635 | 0.36 |
ENST00000522353.2
ENST00000532566.2 |
PCDHA3
|
protocadherin alpha 3 |
| chr1_+_159409512 | 0.36 |
ENST00000423932.3
|
OR10J1
|
olfactory receptor, family 10, subfamily J, member 1 |
| chr11_-_86383650 | 0.36 |
ENST00000526944.1
ENST00000530335.1 ENST00000543262.1 ENST00000524826.1 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr11_-_86383370 | 0.35 |
ENST00000526834.1
ENST00000359636.2 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr1_+_174844645 | 0.35 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr8_-_90996837 | 0.35 |
ENST00000519426.1
ENST00000265433.3 |
NBN
|
nibrin |
| chr11_-_105010320 | 0.34 |
ENST00000532895.1
ENST00000530950.1 |
CARD18
|
caspase recruitment domain family, member 18 |
| chr8_-_42623747 | 0.34 |
ENST00000534622.1
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr7_+_99070527 | 0.34 |
ENST00000379724.3
|
ZNF789
|
zinc finger protein 789 |
| chr1_-_20503917 | 0.33 |
ENST00000429261.2
|
PLA2G2C
|
phospholipase A2, group IIC |
| chr7_-_100253993 | 0.33 |
ENST00000461605.1
ENST00000160382.5 |
ACTL6B
|
actin-like 6B |
| chr15_-_78913628 | 0.33 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr12_+_133757995 | 0.33 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr1_-_182642017 | 0.33 |
ENST00000367557.4
ENST00000258302.4 |
RGS8
|
regulator of G-protein signaling 8 |
| chr14_-_101295407 | 0.33 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr20_+_5731083 | 0.33 |
ENST00000445603.1
ENST00000442185.1 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr12_-_39734783 | 0.33 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr16_-_29517141 | 0.33 |
ENST00000550665.1
|
RP11-231C14.4
|
Uncharacterized protein |
| chr6_-_83775489 | 0.32 |
ENST00000369747.3
|
UBE3D
|
ubiquitin protein ligase E3D |
| chr19_+_17337406 | 0.32 |
ENST00000597836.1
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr2_+_103089756 | 0.32 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr2_-_40680578 | 0.32 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr7_-_92855762 | 0.32 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr5_+_140762268 | 0.31 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr5_-_94417339 | 0.31 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr10_+_118305435 | 0.31 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr11_+_122753391 | 0.31 |
ENST00000307257.6
ENST00000227349.2 |
C11orf63
|
chromosome 11 open reading frame 63 |
| chr18_-_53069419 | 0.31 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr7_-_64023441 | 0.31 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr4_-_120225600 | 0.31 |
ENST00000399075.4
|
C4orf3
|
chromosome 4 open reading frame 3 |
| chr10_+_57358750 | 0.31 |
ENST00000512524.2
|
MTRNR2L5
|
MT-RNR2-like 5 |
| chr3_+_138340067 | 0.30 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr5_-_159827033 | 0.30 |
ENST00000523213.1
|
C5orf54
|
chromosome 5 open reading frame 54 |
| chr8_-_93978346 | 0.30 |
ENST00000523580.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr12_+_8995832 | 0.30 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr19_-_40596828 | 0.30 |
ENST00000414720.2
ENST00000455521.1 ENST00000340963.5 ENST00000595773.1 |
ZNF780A
|
zinc finger protein 780A |
| chr3_-_149510553 | 0.30 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr3_-_151102529 | 0.29 |
ENST00000302632.3
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12 |
| chr12_+_12223867 | 0.29 |
ENST00000308721.5
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr6_-_26108355 | 0.29 |
ENST00000338379.4
|
HIST1H1T
|
histone cluster 1, H1t |
| chr4_-_153601136 | 0.29 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr2_+_53994927 | 0.29 |
ENST00000295304.4
|
CHAC2
|
ChaC, cation transport regulator homolog 2 (E. coli) |
| chr1_+_159750776 | 0.29 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr12_+_74931551 | 0.29 |
ENST00000519948.2
|
ATXN7L3B
|
ataxin 7-like 3B |
| chr8_-_93978357 | 0.29 |
ENST00000522925.1
ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr2_-_208994548 | 0.29 |
ENST00000282141.3
|
CRYGC
|
crystallin, gamma C |
| chr16_+_69796209 | 0.29 |
ENST00000359154.2
ENST00000561780.1 ENST00000563659.1 ENST00000448661.1 |
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr3_+_38347427 | 0.29 |
ENST00000273173.4
|
SLC22A14
|
solute carrier family 22, member 14 |
| chr10_-_69597810 | 0.29 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr9_-_21202204 | 0.28 |
ENST00000239347.3
|
IFNA7
|
interferon, alpha 7 |
| chr16_+_22517166 | 0.28 |
ENST00000356156.3
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr10_+_115614370 | 0.28 |
ENST00000369301.3
|
NHLRC2
|
NHL repeat containing 2 |
| chr8_-_28347737 | 0.28 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr12_-_49418407 | 0.28 |
ENST00000526209.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr4_+_146539415 | 0.28 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr11_+_100862811 | 0.28 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr10_-_123687431 | 0.28 |
ENST00000423243.1
|
ATE1
|
arginyltransferase 1 |
| chr15_+_41549105 | 0.27 |
ENST00000560965.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr1_-_43855444 | 0.27 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr15_-_89089860 | 0.27 |
ENST00000558413.1
ENST00000564406.1 ENST00000268148.8 |
DET1
|
de-etiolated homolog 1 (Arabidopsis) |
| chr3_-_158390282 | 0.27 |
ENST00000264265.3
|
LXN
|
latexin |
| chr2_+_161993465 | 0.27 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr1_+_47489240 | 0.27 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BARX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.5 | 1.4 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.3 | 1.8 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 0.7 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.2 | 0.9 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 0.6 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.2 | 1.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 0.8 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.2 | 0.8 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.2 | 2.0 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.2 | 0.9 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.2 | 3.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.6 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.8 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.5 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.1 | 0.3 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.5 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 4.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.9 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 1.4 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.7 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.1 | 0.2 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.1 | 0.3 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.2 | GO:0051168 | nuclear export(GO:0051168) |
| 0.1 | 0.3 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.2 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.1 | 0.9 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.2 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.6 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 1.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.3 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.5 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.1 | 0.7 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.4 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 0.3 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.2 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.3 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.7 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.1 | 0.3 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.3 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.1 | 0.4 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.6 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.8 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.5 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.3 | GO:1904124 | negative regulation of norepinephrine secretion(GO:0010700) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.2 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.5 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.6 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:1904760 | peptidyl-serine ADP-ribosylation(GO:0018312) myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 1.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.4 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 1.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.7 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.3 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0021940 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.2 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 2.7 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.2 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.2 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.5 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.1 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 2.0 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 1.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.0 | 0.2 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.4 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.1 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.3 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 4.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.3 | GO:0097369 | sodium ion import(GO:0097369) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 2.2 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.3 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.4 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.3 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.0 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 1.3 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.0 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.2 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.5 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.0 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.3 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.3 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.4 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0061458 | reproductive system development(GO:0061458) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.0 | GO:0090290 | hormone-mediated apoptotic signaling pathway(GO:0008628) positive regulation of osteoclast proliferation(GO:0090290) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.3 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.0 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.2 | 0.9 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.2 | 0.5 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 2.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.9 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 2.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 2.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.8 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 1.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 4.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.8 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.0 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 3.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0070469 | mitochondrial respiratory chain(GO:0005746) respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.0 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 1.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 3.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.3 | 1.6 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.3 | 2.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 3.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.2 | 2.0 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.2 | 0.6 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 0.7 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 4.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 0.7 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 2.0 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.4 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.1 | 0.7 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.5 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.1 | 2.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.3 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.6 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 1.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.9 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.2 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.1 | 0.2 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.1 | 0.2 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.2 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 0.4 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 2.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.2 | GO:1904493 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.0 | 0.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.2 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.3 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 1.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 1.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.8 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 3.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.0 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 2.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 0.9 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.8 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |