Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
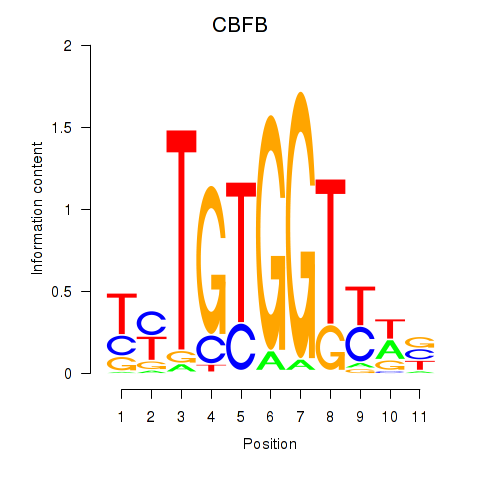
Results for CBFB
Z-value: 0.42
Transcription factors associated with CBFB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CBFB
|
ENSG00000067955.9 | core-binding factor subunit beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CBFB | hg19_v2_chr16_+_67062996_67063019 | 0.27 | 1.5e-01 | Click! |
Activity profile of CBFB motif
Sorted Z-values of CBFB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_44637526 | 1.31 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr14_+_85996471 | 1.07 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr5_-_39203093 | 0.76 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr2_-_158300556 | 0.60 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr19_-_14049184 | 0.60 |
ENST00000339560.5
|
PODNL1
|
podocan-like 1 |
| chr19_-_14048804 | 0.57 |
ENST00000254320.3
ENST00000586075.1 |
PODNL1
|
podocan-like 1 |
| chr14_+_85996507 | 0.55 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr6_-_34524049 | 0.53 |
ENST00000374037.3
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr20_+_62492566 | 0.49 |
ENST00000369916.3
|
ABHD16B
|
abhydrolase domain containing 16B |
| chr6_-_34524093 | 0.46 |
ENST00000544425.1
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr11_-_58343319 | 0.44 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chr3_-_99569821 | 0.37 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr3_+_132843652 | 0.36 |
ENST00000508711.1
|
TMEM108
|
transmembrane protein 108 |
| chr1_-_52499443 | 0.34 |
ENST00000371614.1
|
KTI12
|
KTI12 homolog, chromatin associated (S. cerevisiae) |
| chr20_+_54987305 | 0.33 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr11_-_66103867 | 0.32 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr11_+_114310164 | 0.31 |
ENST00000544196.1
ENST00000539754.1 ENST00000539275.1 |
REXO2
|
RNA exonuclease 2 |
| chr20_+_54987168 | 0.30 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr1_-_179112173 | 0.30 |
ENST00000408940.3
ENST00000504405.1 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr8_-_60031762 | 0.29 |
ENST00000361421.1
|
TOX
|
thymocyte selection-associated high mobility group box |
| chr20_+_43343886 | 0.28 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr11_+_114310102 | 0.26 |
ENST00000265881.5
|
REXO2
|
RNA exonuclease 2 |
| chr12_-_52585765 | 0.25 |
ENST00000313234.5
ENST00000394815.2 |
KRT80
|
keratin 80 |
| chr1_+_47264711 | 0.24 |
ENST00000371923.4
ENST00000271153.4 ENST00000371919.4 |
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1 |
| chr11_-_66104237 | 0.24 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr11_+_114310237 | 0.23 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chrX_+_68048803 | 0.23 |
ENST00000204961.4
|
EFNB1
|
ephrin-B1 |
| chr11_-_66103932 | 0.21 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr17_-_39183452 | 0.21 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr22_+_42665742 | 0.20 |
ENST00000332965.3
ENST00000415205.1 ENST00000446578.1 |
Z83851.3
|
Z83851.3 |
| chr10_-_29811456 | 0.20 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr15_-_64665911 | 0.20 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr12_-_54778471 | 0.20 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chrX_+_115567767 | 0.19 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr4_+_88896819 | 0.19 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr14_+_22748980 | 0.19 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr12_-_14849470 | 0.17 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr12_-_4754339 | 0.17 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr6_+_42847649 | 0.16 |
ENST00000424341.2
ENST00000602561.1 |
RPL7L1
|
ribosomal protein L7-like 1 |
| chr1_+_89246647 | 0.16 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr14_-_59950724 | 0.15 |
ENST00000481608.1
|
L3HYPDH
|
L-3-hydroxyproline dehydratase (trans-) |
| chr14_-_85996332 | 0.15 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr2_+_68592305 | 0.13 |
ENST00000234313.7
|
PLEK
|
pleckstrin |
| chr5_+_131409476 | 0.12 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr14_-_35183886 | 0.12 |
ENST00000298159.6
|
CFL2
|
cofilin 2 (muscle) |
| chr6_+_29068386 | 0.12 |
ENST00000377171.3
|
OR2J1
|
olfactory receptor, family 2, subfamily J, member 1 (gene/pseudogene) |
| chrX_-_49041242 | 0.11 |
ENST00000453382.1
ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr6_-_75912508 | 0.11 |
ENST00000416123.2
|
COL12A1
|
collagen, type XII, alpha 1 |
| chrX_-_70838306 | 0.11 |
ENST00000373691.4
ENST00000373693.3 |
CXCR3
|
chemokine (C-X-C motif) receptor 3 |
| chr4_+_71091786 | 0.10 |
ENST00000317987.5
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr7_+_120628731 | 0.10 |
ENST00000310396.5
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr5_-_137878887 | 0.08 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr3_+_156799587 | 0.08 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr12_-_53594227 | 0.07 |
ENST00000550743.2
|
ITGB7
|
integrin, beta 7 |
| chr3_-_71834318 | 0.06 |
ENST00000353065.3
|
PROK2
|
prokineticin 2 |
| chr15_+_64386261 | 0.06 |
ENST00000560829.1
|
SNX1
|
sorting nexin 1 |
| chr1_+_196621002 | 0.06 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr4_+_74347400 | 0.05 |
ENST00000226355.3
|
AFM
|
afamin |
| chr1_+_39876151 | 0.05 |
ENST00000530275.1
|
KIAA0754
|
KIAA0754 |
| chr2_+_207024306 | 0.05 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr11_+_18343800 | 0.05 |
ENST00000453096.2
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr9_+_15422702 | 0.05 |
ENST00000380821.3
ENST00000421710.1 |
SNAPC3
|
small nuclear RNA activating complex, polypeptide 3, 50kDa |
| chr17_+_32582293 | 0.04 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr6_+_30908747 | 0.04 |
ENST00000462446.1
ENST00000304311.2 |
DPCR1
|
diffuse panbronchiolitis critical region 1 |
| chr6_+_27925019 | 0.04 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr14_+_72064945 | 0.03 |
ENST00000537413.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr1_+_17634689 | 0.03 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr15_+_73976545 | 0.03 |
ENST00000318443.5
ENST00000537340.2 ENST00000318424.5 |
CD276
|
CD276 molecule |
| chr13_-_28674693 | 0.02 |
ENST00000537084.1
ENST00000241453.7 ENST00000380982.4 |
FLT3
|
fms-related tyrosine kinase 3 |
| chr22_+_23222886 | 0.02 |
ENST00000390319.2
|
IGLV3-1
|
immunoglobulin lambda variable 3-1 |
| chrX_-_153718988 | 0.02 |
ENST00000263512.4
ENST00000393587.4 ENST00000453912.1 |
SLC10A3
|
solute carrier family 10, member 3 |
| chr2_+_234826016 | 0.02 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr5_+_35856951 | 0.01 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr8_+_104384616 | 0.01 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr1_-_240775447 | 0.01 |
ENST00000318160.4
|
GREM2
|
gremlin 2, DAN family BMP antagonist |
| chr12_-_55028664 | 0.01 |
ENST00000547511.1
ENST00000257867.4 |
LACRT
|
lacritin |
| chr13_+_76210448 | 0.01 |
ENST00000377499.5
|
LMO7
|
LIM domain 7 |
| chr14_-_59951112 | 0.01 |
ENST00000247194.4
|
L3HYPDH
|
L-3-hydroxyproline dehydratase (trans-) |
| chr2_-_105030466 | 0.01 |
ENST00000449772.1
|
AC068535.3
|
AC068535.3 |
| chr11_-_84028180 | 0.00 |
ENST00000280241.8
|
DLG2
|
discs, large homolog 2 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of CBFB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 1.3 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 1.0 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 0.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.2 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 0.4 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.2 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 1.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.8 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.2 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |