Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
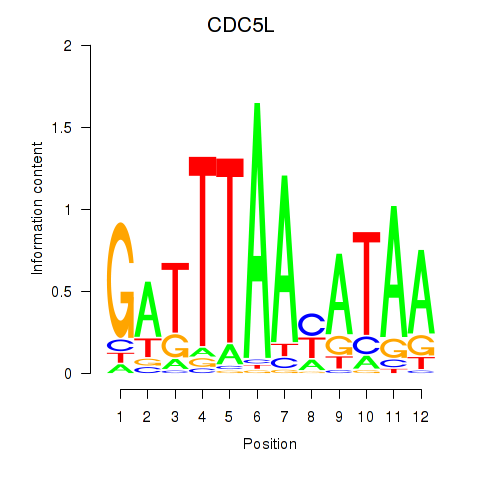
Results for CDC5L
Z-value: 1.06
Transcription factors associated with CDC5L
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDC5L
|
ENSG00000096401.7 | cell division cycle 5 like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDC5L | hg19_v2_chr6_+_44355257_44355315 | 0.11 | 5.8e-01 | Click! |
Activity profile of CDC5L motif
Sorted Z-values of CDC5L motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_16921601 | 7.82 |
ENST00000402239.3
ENST00000310398.2 ENST00000414935.1 |
AGR3
|
anterior gradient 3 |
| chr6_-_32557610 | 6.77 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr16_+_80574854 | 5.13 |
ENST00000305904.6
ENST00000568035.1 |
DYNLRB2
|
dynein, light chain, roadblock-type 2 |
| chr6_-_32498046 | 4.50 |
ENST00000374975.3
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5 |
| chr1_+_38022572 | 4.49 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr5_+_94727048 | 4.08 |
ENST00000283357.5
|
FAM81B
|
family with sequence similarity 81, member B |
| chr5_-_35938674 | 3.73 |
ENST00000397366.1
ENST00000513623.1 ENST00000514524.1 ENST00000397367.2 |
CAPSL
|
calcyphosine-like |
| chr1_+_38022513 | 3.68 |
ENST00000296218.7
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr6_+_131958436 | 3.56 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr4_-_100356844 | 3.43 |
ENST00000437033.2
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr6_-_33041378 | 3.29 |
ENST00000428995.1
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr17_+_45908974 | 3.29 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr19_-_9006766 | 3.09 |
ENST00000599436.1
|
MUC16
|
mucin 16, cell surface associated |
| chr10_+_115511213 | 3.03 |
ENST00000361048.1
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr11_+_73661364 | 2.88 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr12_+_20963647 | 2.80 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr12_+_20963632 | 2.74 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr13_+_50589390 | 2.69 |
ENST00000360473.4
ENST00000312942.1 |
KCNRG
|
potassium channel regulator |
| chr1_-_150738261 | 2.44 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr6_+_32709119 | 2.42 |
ENST00000374940.3
|
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 2 |
| chr1_-_104238912 | 2.41 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr1_-_104239076 | 2.36 |
ENST00000370080.3
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr5_+_140227048 | 2.35 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr2_+_102721023 | 2.32 |
ENST00000409589.1
ENST00000409329.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr3_-_120365866 | 2.19 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr3_+_160559931 | 2.17 |
ENST00000464260.1
ENST00000295839.9 |
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr4_-_70725856 | 2.15 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr5_+_140213815 | 2.06 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr12_+_21168630 | 2.03 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr2_-_190445499 | 1.96 |
ENST00000261024.2
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr6_-_165723088 | 1.82 |
ENST00000230301.8
|
C6orf118
|
chromosome 6 open reading frame 118 |
| chr18_-_24765248 | 1.79 |
ENST00000580774.1
ENST00000284224.8 |
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr12_+_20968608 | 1.78 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr8_-_133637624 | 1.75 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr6_-_39197226 | 1.71 |
ENST00000359534.3
|
KCNK5
|
potassium channel, subfamily K, member 5 |
| chr12_+_21207503 | 1.69 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr8_+_76452097 | 1.65 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr4_-_100356291 | 1.64 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr10_-_61495760 | 1.57 |
ENST00000395347.1
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr10_+_51549498 | 1.56 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr5_+_140220769 | 1.53 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chr10_+_118305435 | 1.51 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr3_+_108321623 | 1.51 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr7_+_116654935 | 1.50 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chrX_+_36246735 | 1.48 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr11_-_111944704 | 1.47 |
ENST00000532211.1
|
PIH1D2
|
PIH1 domain containing 2 |
| chr1_+_144339738 | 1.46 |
ENST00000538264.1
|
AL592284.1
|
Protein LOC642441 |
| chr1_+_59775752 | 1.46 |
ENST00000371212.1
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr6_-_52705641 | 1.45 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr4_-_100356551 | 1.44 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr8_-_54436491 | 1.44 |
ENST00000426023.1
|
RP11-400K9.4
|
RP11-400K9.4 |
| chr10_-_62493223 | 1.40 |
ENST00000373827.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr4_-_38806404 | 1.39 |
ENST00000308979.2
ENST00000505940.1 ENST00000515861.1 |
TLR1
|
toll-like receptor 1 |
| chr5_+_140165876 | 1.38 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr4_+_41614909 | 1.37 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_+_156338993 | 1.35 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr10_-_69597915 | 1.34 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr13_-_86373536 | 1.34 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr11_-_111944895 | 1.31 |
ENST00000431456.1
ENST00000280350.4 ENST00000530641.1 |
PIH1D2
|
PIH1 domain containing 2 |
| chr1_-_86848760 | 1.25 |
ENST00000460698.2
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr10_-_116286563 | 1.24 |
ENST00000369253.2
|
ABLIM1
|
actin binding LIM protein 1 |
| chr6_+_32812568 | 1.21 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr21_+_33784670 | 1.21 |
ENST00000300255.2
|
EVA1C
|
eva-1 homolog C (C. elegans) |
| chrX_+_36254051 | 1.20 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr12_+_133757995 | 1.19 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr4_+_41614720 | 1.19 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr12_+_9980113 | 1.18 |
ENST00000537723.1
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr2_+_232573208 | 1.14 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr11_-_85430163 | 1.13 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr14_-_21516590 | 1.12 |
ENST00000555026.1
|
NDRG2
|
NDRG family member 2 |
| chr19_-_11545920 | 1.12 |
ENST00000356392.4
ENST00000591179.1 |
CCDC151
|
coiled-coil domain containing 151 |
| chr5_+_140514782 | 1.10 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr4_-_141348999 | 1.07 |
ENST00000325617.5
|
CLGN
|
calmegin |
| chr5_+_140180635 | 1.02 |
ENST00000522353.2
ENST00000532566.2 |
PCDHA3
|
protocadherin alpha 3 |
| chr5_-_61031495 | 1.01 |
ENST00000506550.1
ENST00000512882.2 |
CTD-2170G1.2
|
CTD-2170G1.2 |
| chr8_+_104831554 | 1.00 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr2_+_232573222 | 0.99 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr2_-_222436988 | 0.98 |
ENST00000409854.1
ENST00000281821.2 ENST00000392071.4 ENST00000443796.1 |
EPHA4
|
EPH receptor A4 |
| chr17_+_37856299 | 0.97 |
ENST00000269571.5
|
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr19_-_49843539 | 0.97 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr9_+_90112117 | 0.96 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr10_-_116444371 | 0.96 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr7_+_116593433 | 0.96 |
ENST00000323984.3
ENST00000393449.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr1_-_86043921 | 0.95 |
ENST00000535924.2
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr12_-_39734783 | 0.94 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr17_+_37856253 | 0.90 |
ENST00000540147.1
ENST00000584450.1 |
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr5_-_179499108 | 0.89 |
ENST00000521389.1
|
RNF130
|
ring finger protein 130 |
| chr14_-_106453155 | 0.89 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr8_+_21911054 | 0.88 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr5_-_179499086 | 0.88 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130 |
| chr11_-_85430088 | 0.85 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr17_+_37856214 | 0.85 |
ENST00000445658.2
|
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr4_-_48082192 | 0.84 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr5_-_111093759 | 0.84 |
ENST00000509979.1
ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP
|
neuronal regeneration related protein |
| chr6_+_33422343 | 0.82 |
ENST00000395064.2
|
ZBTB9
|
zinc finger and BTB domain containing 9 |
| chr22_+_29168652 | 0.81 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr10_-_69597810 | 0.80 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr10_+_96698406 | 0.80 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr5_+_139505520 | 0.79 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr16_+_78056412 | 0.79 |
ENST00000299642.4
ENST00000575655.1 |
CLEC3A
|
C-type lectin domain family 3, member A |
| chr10_-_1246317 | 0.78 |
ENST00000381305.1
|
ADARB2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr1_+_196621156 | 0.77 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr5_+_147691979 | 0.77 |
ENST00000274565.4
|
SPINK7
|
serine peptidase inhibitor, Kazal type 7 (putative) |
| chr12_+_133758115 | 0.77 |
ENST00000541009.2
ENST00000592241.1 |
ZNF268
|
zinc finger protein 268 |
| chr17_+_7788104 | 0.77 |
ENST00000380358.4
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr10_-_28623368 | 0.77 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr12_-_111358372 | 0.77 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr12_+_12223867 | 0.75 |
ENST00000308721.5
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr11_-_114466477 | 0.74 |
ENST00000375478.3
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr17_-_73775839 | 0.73 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chrX_+_35937843 | 0.73 |
ENST00000297866.5
|
CXorf22
|
chromosome X open reading frame 22 |
| chr14_+_67291158 | 0.72 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr13_-_41768654 | 0.71 |
ENST00000379483.3
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr1_-_198990166 | 0.71 |
ENST00000427439.1
|
RP11-16L9.3
|
RP11-16L9.3 |
| chr3_-_155394099 | 0.70 |
ENST00000414191.1
|
PLCH1
|
phospholipase C, eta 1 |
| chr3_-_197300194 | 0.69 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr4_+_37455536 | 0.65 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr8_+_19171128 | 0.64 |
ENST00000265807.3
|
SH2D4A
|
SH2 domain containing 4A |
| chr11_+_124055923 | 0.64 |
ENST00000318666.6
|
OR10D3
|
olfactory receptor, family 10, subfamily D, member 3 (non-functional) |
| chr4_+_41540160 | 0.64 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_-_127541194 | 0.64 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr21_+_43619796 | 0.63 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr19_+_35939154 | 0.63 |
ENST00000599180.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr6_+_27791862 | 0.62 |
ENST00000355057.1
|
HIST1H4J
|
histone cluster 1, H4j |
| chr2_-_110962544 | 0.62 |
ENST00000355301.4
ENST00000445609.2 ENST00000417665.1 ENST00000418527.1 ENST00000316534.4 ENST00000393272.3 |
NPHP1
|
nephronophthisis 1 (juvenile) |
| chr15_-_43876702 | 0.61 |
ENST00000348806.6
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr1_+_174769006 | 0.61 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr20_-_18477862 | 0.61 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr16_+_4845379 | 0.61 |
ENST00000588606.1
ENST00000586005.1 |
SMIM22
|
small integral membrane protein 22 |
| chrM_+_12331 | 0.60 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr12_+_55248289 | 0.60 |
ENST00000308796.6
|
MUCL1
|
mucin-like 1 |
| chr8_+_99956759 | 0.60 |
ENST00000522510.1
ENST00000457907.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr4_+_170541660 | 0.59 |
ENST00000513761.1
ENST00000347613.4 |
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr1_-_216978709 | 0.59 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr9_-_115653176 | 0.58 |
ENST00000374228.4
|
SLC46A2
|
solute carrier family 46, member 2 |
| chr1_+_166958346 | 0.58 |
ENST00000367872.4
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr12_-_25150409 | 0.58 |
ENST00000549262.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr4_-_69111401 | 0.58 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr4_-_87028478 | 0.58 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr3_+_171561127 | 0.57 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr10_+_106113515 | 0.57 |
ENST00000369704.3
ENST00000312902.5 |
CCDC147
|
coiled-coil domain containing 147 |
| chr19_-_10697895 | 0.56 |
ENST00000591240.1
ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chrX_-_131262048 | 0.56 |
ENST00000298542.4
|
FRMD7
|
FERM domain containing 7 |
| chr16_+_57576584 | 0.56 |
ENST00000340339.4
|
GPR114
|
G protein-coupled receptor 114 |
| chr10_+_111765562 | 0.56 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr6_-_76072719 | 0.55 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr12_+_80603233 | 0.55 |
ENST00000547103.1
ENST00000458043.2 |
OTOGL
|
otogelin-like |
| chr15_+_36338242 | 0.55 |
ENST00000560056.1
|
RP11-684B21.1
|
RP11-684B21.1 |
| chr1_+_152635854 | 0.55 |
ENST00000368784.1
|
LCE2D
|
late cornified envelope 2D |
| chr17_-_35969409 | 0.54 |
ENST00000394378.2
ENST00000585472.1 ENST00000591288.1 ENST00000502449.2 ENST00000345615.4 ENST00000346661.4 ENST00000585689.1 ENST00000339208.6 |
SYNRG
|
synergin, gamma |
| chr12_-_65146636 | 0.54 |
ENST00000418919.2
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr4_+_130017268 | 0.53 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr14_+_76452090 | 0.53 |
ENST00000314067.6
ENST00000238628.6 ENST00000556742.1 |
IFT43
|
intraflagellar transport 43 homolog (Chlamydomonas) |
| chrX_+_144908928 | 0.52 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr2_+_66662510 | 0.52 |
ENST00000272369.9
ENST00000407092.2 |
MEIS1
|
Meis homeobox 1 |
| chr1_+_9599540 | 0.52 |
ENST00000302692.6
|
SLC25A33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr21_-_43816052 | 0.51 |
ENST00000398405.1
|
TMPRSS3
|
transmembrane protease, serine 3 |
| chr6_-_49681235 | 0.50 |
ENST00000339139.4
|
CRISP2
|
cysteine-rich secretory protein 2 |
| chr16_-_5116025 | 0.50 |
ENST00000472572.3
ENST00000315997.5 ENST00000422873.1 ENST00000350219.4 |
C16orf89
|
chromosome 16 open reading frame 89 |
| chr2_-_160473114 | 0.50 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr1_-_89591749 | 0.50 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr8_-_57233103 | 0.50 |
ENST00000303749.3
ENST00000522671.1 |
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5 |
| chr14_+_62585332 | 0.49 |
ENST00000554895.1
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
| chr11_-_31531121 | 0.49 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr5_-_10761206 | 0.49 |
ENST00000432074.2
ENST00000230895.6 |
DAP
|
death-associated protein |
| chr5_+_8839844 | 0.49 |
ENST00000510229.1
ENST00000506655.1 ENST00000510067.1 |
RP11-143A12.3
|
RP11-143A12.3 |
| chr1_+_41204506 | 0.48 |
ENST00000525290.1
ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr2_+_127413481 | 0.48 |
ENST00000259254.4
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr15_+_65822756 | 0.48 |
ENST00000562901.1
ENST00000261875.5 ENST00000442729.2 ENST00000565299.1 ENST00000568793.1 |
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr1_-_76076793 | 0.48 |
ENST00000370859.3
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr1_+_70820451 | 0.48 |
ENST00000361764.4
ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3
|
HERV-H LTR-associating 3 |
| chr1_-_93257951 | 0.47 |
ENST00000543509.1
ENST00000370331.1 ENST00000540033.1 |
EVI5
|
ecotropic viral integration site 5 |
| chr1_+_22351977 | 0.47 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr6_+_80129989 | 0.47 |
ENST00000429444.1
|
RP1-232L24.3
|
RP1-232L24.3 |
| chr19_-_48752812 | 0.46 |
ENST00000359009.4
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr11_-_236326 | 0.45 |
ENST00000525237.1
ENST00000532956.1 ENST00000525319.1 ENST00000524564.1 ENST00000382743.4 |
SIRT3
|
sirtuin 3 |
| chr3_-_93747425 | 0.45 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr8_+_104892639 | 0.45 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr6_-_119031228 | 0.44 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr2_+_54350316 | 0.44 |
ENST00000606865.1
|
ACYP2
|
acylphosphatase 2, muscle type |
| chr8_+_7353368 | 0.44 |
ENST00000355602.2
|
DEFB107B
|
defensin, beta 107B |
| chr3_+_108541545 | 0.43 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr15_+_65823092 | 0.43 |
ENST00000566074.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr3_+_69928256 | 0.43 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chrX_-_131228291 | 0.42 |
ENST00000370879.1
|
FRMD7
|
FERM domain containing 7 |
| chr13_+_111855414 | 0.41 |
ENST00000375737.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr6_+_144904334 | 0.41 |
ENST00000367526.4
|
UTRN
|
utrophin |
| chr15_-_77376269 | 0.41 |
ENST00000558745.1
|
TSPAN3
|
tetraspanin 3 |
| chr1_-_153283194 | 0.41 |
ENST00000290722.1
|
PGLYRP3
|
peptidoglycan recognition protein 3 |
| chr3_+_20081515 | 0.40 |
ENST00000263754.4
|
KAT2B
|
K(lysine) acetyltransferase 2B |
| chr10_+_81462983 | 0.40 |
ENST00000448135.1
ENST00000429828.1 ENST00000372321.1 |
NUTM2B
|
NUT family member 2B |
| chr2_+_233527443 | 0.40 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr19_-_4540486 | 0.40 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr17_+_4336955 | 0.40 |
ENST00000355530.2
|
SPNS3
|
spinster homolog 3 (Drosophila) |
| chr3_+_108541608 | 0.39 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr19_-_48753104 | 0.39 |
ENST00000447740.2
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr22_-_39268192 | 0.39 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr5_-_157286104 | 0.38 |
ENST00000530742.1
ENST00000523908.1 ENST00000523094.1 ENST00000296951.5 ENST00000411809.2 |
CLINT1
|
clathrin interactor 1 |
| chr1_+_196621002 | 0.38 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr10_-_13390270 | 0.38 |
ENST00000378614.4
ENST00000545675.1 ENST00000327347.5 |
SEPHS1
|
selenophosphate synthetase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CDC5L
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 1.4 | 6.8 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 1.2 | 3.6 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.8 | 2.3 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.7 | 2.2 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.7 | 2.0 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.7 | 2.0 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.6 | 2.9 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.5 | 1.4 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.4 | 1.1 | GO:0070340 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.4 | 1.4 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 | 2.7 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.3 | 1.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.3 | 2.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.7 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.2 | 1.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 10.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 0.6 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.2 | 2.7 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.2 | 0.6 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 0.9 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.2 | 0.4 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.2 | 1.4 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 1.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.2 | 0.5 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 1.7 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 0.6 | GO:0055099 | detection of endogenous stimulus(GO:0009726) response to high density lipoprotein particle(GO:0055099) |
| 0.2 | 2.0 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.2 | 1.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.1 | 0.9 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.8 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.6 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 0.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.4 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.4 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 5.9 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.4 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 | 0.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 2.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.3 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.8 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.4 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 1.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 1.3 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.1 | 1.5 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 1.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.8 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.3 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 10.7 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 1.6 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 | 1.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.8 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.3 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.2 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.3 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 | 0.3 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.5 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 0.2 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.2 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.6 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.3 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.0 | 1.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 3.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 7.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.4 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 3.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.6 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.2 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 1.0 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 3.4 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.5 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |
| 0.0 | 0.5 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.6 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.3 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.3 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.2 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 2.1 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 1.6 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.3 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.1 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.1 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.4 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.0 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.0 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.4 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.3 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:0045137 | gonad development(GO:0008406) development of primary sexual characteristics(GO:0045137) |
| 0.0 | 1.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.5 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 17.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.5 | 5.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.4 | 1.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.3 | 2.4 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.2 | 1.5 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 0.6 | GO:0030849 | autosome(GO:0030849) |
| 0.2 | 3.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 0.5 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 8.4 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 1.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.8 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.4 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.1 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 3.2 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 2.1 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 10.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.7 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 5.0 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.5 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.7 | 3.6 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.6 | 2.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.5 | 10.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.4 | 5.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.4 | 8.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.4 | 11.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.3 | 2.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 1.8 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.3 | 1.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.2 | 0.7 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.2 | 0.7 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.2 | 2.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 0.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 2.0 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 0.6 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.2 | 5.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 5.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.2 | 1.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 1.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 0.5 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 0.8 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.6 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.8 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.4 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.6 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.4 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 1.5 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 0.4 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 0.6 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 2.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 1.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) PH domain binding(GO:0042731) |
| 0.1 | 2.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 3.6 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 0.3 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.6 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 1.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.2 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.1 | 0.4 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.2 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 2.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.0 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.5 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 1.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 1.5 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 1.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 1.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 2.9 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.3 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 1.3 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.3 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 1.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 3.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.6 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.1 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.4 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 16.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 4.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.5 | 6.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 6.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 2.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 3.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 2.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 1.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.8 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.8 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.2 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |