Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
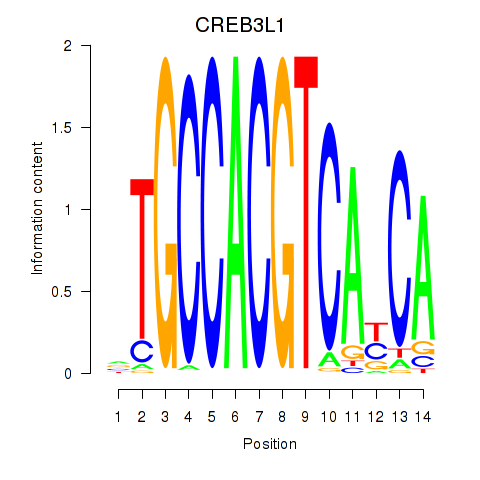
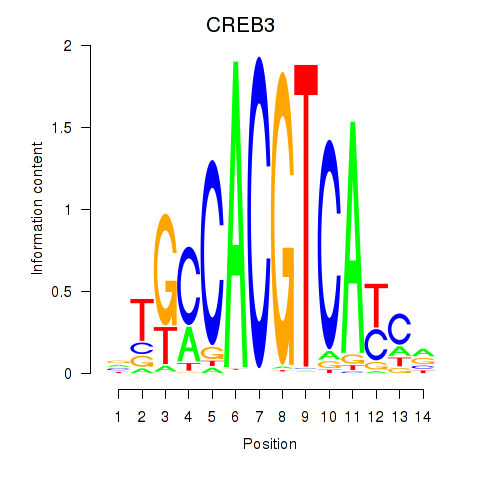
Results for CREB3L1_CREB3
Z-value: 0.64
Transcription factors associated with CREB3L1_CREB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L1
|
ENSG00000157613.6 | cAMP responsive element binding protein 3 like 1 |
|
CREB3
|
ENSG00000107175.6 | cAMP responsive element binding protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB3 | hg19_v2_chr9_+_35732312_35732332 | -0.30 | 1.0e-01 | Click! |
| CREB3L1 | hg19_v2_chr11_+_46299199_46299233 | -0.24 | 2.1e-01 | Click! |
Activity profile of CREB3L1_CREB3 motif
Sorted Z-values of CREB3L1_CREB3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_113498616 | 2.17 |
ENST00000433570.4
ENST00000538576.1 ENST00000458229.1 |
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr1_-_113498943 | 2.14 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr4_+_75311019 | 1.84 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr4_+_75310851 | 1.74 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr12_+_41086297 | 1.58 |
ENST00000551295.2
|
CNTN1
|
contactin 1 |
| chr9_+_124461603 | 1.46 |
ENST00000373782.3
|
DAB2IP
|
DAB2 interacting protein |
| chr6_+_151561085 | 1.28 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr11_-_64646086 | 1.27 |
ENST00000320631.3
|
EHD1
|
EH-domain containing 1 |
| chr4_-_122744998 | 1.13 |
ENST00000274026.5
|
CCNA2
|
cyclin A2 |
| chr19_+_38810447 | 1.11 |
ENST00000263372.3
|
KCNK6
|
potassium channel, subfamily K, member 6 |
| chr11_+_125495862 | 1.08 |
ENST00000428830.2
ENST00000544373.1 ENST00000527013.1 ENST00000526937.1 ENST00000534685.1 |
CHEK1
|
checkpoint kinase 1 |
| chr3_-_156272924 | 1.07 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr5_+_94890840 | 0.98 |
ENST00000504763.1
|
ARSK
|
arylsulfatase family, member K |
| chr14_+_68086515 | 0.92 |
ENST00000261783.3
|
ARG2
|
arginase 2 |
| chr1_+_45205478 | 0.90 |
ENST00000452259.1
ENST00000372224.4 |
KIF2C
|
kinesin family member 2C |
| chr22_+_38864041 | 0.90 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr1_+_45205498 | 0.89 |
ENST00000372218.4
|
KIF2C
|
kinesin family member 2C |
| chr19_+_41256764 | 0.86 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr3_+_100211412 | 0.83 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr7_+_116593953 | 0.82 |
ENST00000397750.3
|
ST7-OT4
|
ST7 overlapping transcript 4 |
| chr19_-_49015050 | 0.75 |
ENST00000600059.1
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr5_-_94890648 | 0.74 |
ENST00000513823.1
ENST00000514952.1 ENST00000358746.2 |
TTC37
|
tetratricopeptide repeat domain 37 |
| chr3_-_156272872 | 0.72 |
ENST00000476217.1
|
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr1_-_119682812 | 0.70 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr12_-_113909877 | 0.69 |
ENST00000261731.3
|
LHX5
|
LIM homeobox 5 |
| chr6_+_24775641 | 0.68 |
ENST00000378054.2
ENST00000476555.1 |
GMNN
|
geminin, DNA replication inhibitor |
| chr20_-_1306351 | 0.62 |
ENST00000381812.1
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr8_+_22423219 | 0.62 |
ENST00000523965.1
ENST00000521554.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr7_-_30066233 | 0.61 |
ENST00000222803.5
|
FKBP14
|
FK506 binding protein 14, 22 kDa |
| chr11_+_125496124 | 0.60 |
ENST00000533778.2
ENST00000534070.1 |
CHEK1
|
checkpoint kinase 1 |
| chr19_-_4670345 | 0.60 |
ENST00000599630.1
ENST00000262947.3 |
C19orf10
|
chromosome 19 open reading frame 10 |
| chr17_+_7211656 | 0.59 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr3_-_121468513 | 0.59 |
ENST00000494517.1
ENST00000393667.3 |
GOLGB1
|
golgin B1 |
| chr20_-_1306391 | 0.59 |
ENST00000339987.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr2_-_202645612 | 0.59 |
ENST00000409632.2
ENST00000410052.1 ENST00000467448.1 |
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr15_+_73976715 | 0.58 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr10_-_71930222 | 0.58 |
ENST00000458634.2
ENST00000373239.2 ENST00000373242.2 ENST00000373241.4 |
SAR1A
|
SAR1 homolog A (S. cerevisiae) |
| chr6_+_24775153 | 0.58 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr2_+_183580954 | 0.57 |
ENST00000264065.7
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr3_-_57583185 | 0.57 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4 |
| chr3_-_121468602 | 0.57 |
ENST00000340645.5
|
GOLGB1
|
golgin B1 |
| chr11_+_69455855 | 0.57 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr9_+_112542572 | 0.56 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough |
| chr12_-_106641728 | 0.55 |
ENST00000378026.4
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr3_+_50284321 | 0.50 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr6_-_7313381 | 0.50 |
ENST00000489567.1
ENST00000479365.1 ENST00000462112.1 ENST00000397511.2 ENST00000534851.1 ENST00000474597.1 ENST00000244763.4 |
SSR1
|
signal sequence receptor, alpha |
| chr19_+_39903185 | 0.50 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr8_+_22422749 | 0.50 |
ENST00000523900.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr3_-_28390298 | 0.50 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr19_-_41196534 | 0.50 |
ENST00000252891.4
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr20_-_35402123 | 0.49 |
ENST00000373740.3
ENST00000426836.1 ENST00000373745.3 ENST00000448110.2 ENST00000438549.1 ENST00000447406.1 ENST00000373750.4 ENST00000373734.4 |
DSN1
|
DSN1, MIS12 kinetochore complex component |
| chr19_+_5681153 | 0.48 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr14_+_24600484 | 0.47 |
ENST00000267426.5
|
FITM1
|
fat storage-inducing transmembrane protein 1 |
| chr1_-_119683251 | 0.46 |
ENST00000369426.5
ENST00000235521.4 |
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr5_-_114961673 | 0.46 |
ENST00000333314.3
|
TMED7-TICAM2
|
TMED7-TICAM2 readthrough |
| chr5_-_131562935 | 0.45 |
ENST00000379104.2
ENST00000379100.2 ENST00000428369.1 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr11_-_67407031 | 0.44 |
ENST00000335385.3
|
TBX10
|
T-box 10 |
| chr5_-_131563501 | 0.44 |
ENST00000401867.1
ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr6_+_116692102 | 0.42 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase |
| chr11_-_2906979 | 0.42 |
ENST00000380725.1
ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chrX_+_47441712 | 0.41 |
ENST00000218388.4
ENST00000377018.2 ENST00000456754.2 ENST00000377017.1 ENST00000441738.1 |
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr7_+_128379449 | 0.40 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr16_-_10868853 | 0.39 |
ENST00000572428.1
|
TVP23A
|
trans-golgi network vesicle protein 23 homolog A (S. cerevisiae) |
| chr17_+_48450575 | 0.39 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr12_-_122750957 | 0.39 |
ENST00000451053.2
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae) |
| chr21_-_34960930 | 0.38 |
ENST00000437395.1
|
DONSON
|
downstream neighbor of SON |
| chr19_+_16187816 | 0.38 |
ENST00000588410.1
|
TPM4
|
tropomyosin 4 |
| chr3_-_28390415 | 0.38 |
ENST00000414162.1
ENST00000420543.2 |
AZI2
|
5-azacytidine induced 2 |
| chr11_+_65686728 | 0.37 |
ENST00000312515.2
ENST00000525501.1 |
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr11_+_65686802 | 0.36 |
ENST00000376991.2
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr19_-_5680891 | 0.36 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr17_-_7137857 | 0.36 |
ENST00000005340.5
|
DVL2
|
dishevelled segment polarity protein 2 |
| chr8_-_29208183 | 0.36 |
ENST00000240100.2
|
DUSP4
|
dual specificity phosphatase 4 |
| chr19_+_49458107 | 0.35 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
| chr22_+_46476192 | 0.35 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chr10_+_27444268 | 0.35 |
ENST00000375940.4
ENST00000342386.6 |
MASTL
|
microtubule associated serine/threonine kinase-like |
| chr11_-_67276100 | 0.34 |
ENST00000301488.3
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr7_-_100860851 | 0.33 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr17_-_7137582 | 0.32 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr9_+_101984577 | 0.32 |
ENST00000223641.4
|
SEC61B
|
Sec61 beta subunit |
| chr15_+_26360970 | 0.32 |
ENST00000556159.1
ENST00000557523.1 |
LINC00929
|
long intergenic non-protein coding RNA 929 |
| chr5_+_133984462 | 0.32 |
ENST00000398844.2
ENST00000322887.4 |
SEC24A
|
SEC24 family member A |
| chr1_-_11865982 | 0.32 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr10_+_26932068 | 0.32 |
ENST00000544033.1
ENST00000454991.1 |
LINC00202-2
RP13-16H11.7
|
long intergenic non-protein coding RNA 202-2 RP13-16H11.7 |
| chr3_-_28390120 | 0.31 |
ENST00000334100.6
|
AZI2
|
5-azacytidine induced 2 |
| chr13_+_113951607 | 0.30 |
ENST00000397181.3
|
LAMP1
|
lysosomal-associated membrane protein 1 |
| chr5_+_176730769 | 0.30 |
ENST00000303204.4
ENST00000503216.1 |
PRELID1
|
PRELI domain containing 1 |
| chr17_-_7155274 | 0.30 |
ENST00000318988.6
ENST00000575783.1 ENST00000573600.1 |
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr16_+_81040794 | 0.30 |
ENST00000439957.3
ENST00000393335.3 ENST00000428963.2 ENST00000564669.1 |
CENPN
|
centromere protein N |
| chr4_+_141445311 | 0.30 |
ENST00000323570.3
ENST00000511887.2 |
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr12_+_48722763 | 0.30 |
ENST00000335017.1
|
H1FNT
|
H1 histone family, member N, testis-specific |
| chrX_-_46618490 | 0.30 |
ENST00000328306.4
|
SLC9A7
|
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chr2_+_192542850 | 0.29 |
ENST00000410026.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr19_-_41196458 | 0.28 |
ENST00000598779.1
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr3_+_133292574 | 0.28 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chr1_-_160313025 | 0.28 |
ENST00000368069.3
ENST00000241704.7 |
COPA
|
coatomer protein complex, subunit alpha |
| chr6_-_151773232 | 0.28 |
ENST00000444024.1
ENST00000367303.4 |
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr1_+_44445549 | 0.28 |
ENST00000356836.6
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr5_+_68389807 | 0.28 |
ENST00000380860.4
ENST00000504103.1 ENST00000502979.1 |
SLC30A5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr18_-_46987000 | 0.28 |
ENST00000442713.2
ENST00000269445.6 |
DYM
|
dymeclin |
| chr8_+_22423168 | 0.28 |
ENST00000518912.1
ENST00000428103.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr22_+_32754139 | 0.27 |
ENST00000382088.3
|
RFPL3
|
ret finger protein-like 3 |
| chr3_-_57583130 | 0.26 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr4_-_119757239 | 0.26 |
ENST00000280551.6
|
SEC24D
|
SEC24 family member D |
| chr21_+_45138941 | 0.26 |
ENST00000398081.1
ENST00000468090.1 ENST00000291565.4 |
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr17_-_46716647 | 0.25 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr19_-_48894104 | 0.25 |
ENST00000597017.1
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chrX_-_7895755 | 0.25 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr17_-_7154984 | 0.25 |
ENST00000574322.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr14_+_78227105 | 0.25 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr15_+_40987327 | 0.25 |
ENST00000423169.2
ENST00000267868.3 ENST00000557850.1 ENST00000532743.1 ENST00000382643.3 |
RAD51
|
RAD51 recombinase |
| chr21_+_44313375 | 0.24 |
ENST00000354250.2
ENST00000340344.4 |
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr9_+_126777676 | 0.24 |
ENST00000488674.2
|
LHX2
|
LIM homeobox 2 |
| chr19_-_47164386 | 0.24 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr12_+_113376249 | 0.24 |
ENST00000551007.1
ENST00000548514.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr19_-_46916805 | 0.23 |
ENST00000307522.3
|
CCDC8
|
coiled-coil domain containing 8 |
| chr17_+_40688190 | 0.23 |
ENST00000225927.2
|
NAGLU
|
N-acetylglucosaminidase, alpha |
| chr15_-_23692381 | 0.23 |
ENST00000567107.1
ENST00000345070.5 ENST00000312015.5 |
GOLGA6L2
|
golgin A6 family-like 2 |
| chr1_-_43833628 | 0.23 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr1_-_11866034 | 0.23 |
ENST00000376590.3
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr1_+_26758790 | 0.23 |
ENST00000427245.2
ENST00000525682.2 ENST00000236342.7 ENST00000526219.1 ENST00000374185.3 ENST00000360009.2 |
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr3_-_113465065 | 0.23 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr3_+_122785895 | 0.22 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr22_+_32753854 | 0.22 |
ENST00000249007.4
|
RFPL3
|
ret finger protein-like 3 |
| chr2_-_220042825 | 0.22 |
ENST00000409789.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr15_-_40213080 | 0.22 |
ENST00000561100.1
|
GPR176
|
G protein-coupled receptor 176 |
| chr13_+_113951532 | 0.22 |
ENST00000332556.4
|
LAMP1
|
lysosomal-associated membrane protein 1 |
| chr2_+_219745020 | 0.22 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr5_-_114961858 | 0.22 |
ENST00000282382.4
ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2
TMED7
TICAM2
|
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr16_+_56642041 | 0.22 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr10_+_64893039 | 0.22 |
ENST00000277746.6
ENST00000435510.2 |
NRBF2
|
nuclear receptor binding factor 2 |
| chr5_-_39425290 | 0.22 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr16_-_2059748 | 0.22 |
ENST00000562103.1
ENST00000431526.1 |
ZNF598
|
zinc finger protein 598 |
| chr7_+_39663061 | 0.21 |
ENST00000005257.2
|
RALA
|
v-ral simian leukemia viral oncogene homolog A (ras related) |
| chr11_+_66188475 | 0.21 |
ENST00000311034.2
|
NPAS4
|
neuronal PAS domain protein 4 |
| chr2_-_80531399 | 0.21 |
ENST00000409148.1
ENST00000415098.1 ENST00000452811.1 |
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr5_+_94890778 | 0.21 |
ENST00000380009.4
|
ARSK
|
arylsulfatase family, member K |
| chr2_-_130886795 | 0.21 |
ENST00000409914.2
|
POTEF
|
POTE ankyrin domain family, member F |
| chr8_-_144623595 | 0.21 |
ENST00000262577.5
|
ZC3H3
|
zinc finger CCCH-type containing 3 |
| chr8_-_57906362 | 0.21 |
ENST00000262644.4
|
IMPAD1
|
inositol monophosphatase domain containing 1 |
| chr5_-_138210977 | 0.21 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr6_-_101329157 | 0.21 |
ENST00000369143.2
|
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr16_+_56642489 | 0.21 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A |
| chr2_-_80531824 | 0.21 |
ENST00000295057.3
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr2_+_28615669 | 0.21 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr1_-_202311088 | 0.21 |
ENST00000367274.4
|
UBE2T
|
ubiquitin-conjugating enzyme E2T (putative) |
| chr4_-_83350580 | 0.20 |
ENST00000349655.4
ENST00000602300.1 |
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr19_+_531713 | 0.20 |
ENST00000215574.4
|
CDC34
|
cell division cycle 34 |
| chr1_+_55271736 | 0.20 |
ENST00000358193.3
ENST00000371273.3 |
C1orf177
|
chromosome 1 open reading frame 177 |
| chr19_-_18391708 | 0.20 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr9_-_136242956 | 0.20 |
ENST00000371989.3
ENST00000485435.2 |
SURF4
|
surfeit 4 |
| chr5_-_39425222 | 0.19 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr9_+_131709966 | 0.19 |
ENST00000372577.2
|
NUP188
|
nucleoporin 188kDa |
| chr5_+_109025067 | 0.19 |
ENST00000261483.4
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1 |
| chr17_-_6554877 | 0.19 |
ENST00000225728.3
ENST00000575197.1 |
MED31
|
mediator complex subunit 31 |
| chr5_-_172198190 | 0.19 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr6_-_90062543 | 0.18 |
ENST00000435041.2
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr7_+_128379346 | 0.18 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr16_+_2059872 | 0.18 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr22_+_21271714 | 0.18 |
ENST00000354336.3
|
CRKL
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr12_-_50616382 | 0.18 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr19_-_15236173 | 0.18 |
ENST00000527093.1
|
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr19_+_797392 | 0.17 |
ENST00000350092.4
ENST00000349038.4 ENST00000586481.1 ENST00000585535.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr1_-_32110467 | 0.17 |
ENST00000440872.2
ENST00000373703.4 |
PEF1
|
penta-EF-hand domain containing 1 |
| chr19_-_10341948 | 0.17 |
ENST00000590320.1
ENST00000592342.1 ENST00000588952.1 |
S1PR2
DNMT1
|
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr15_+_84904525 | 0.17 |
ENST00000510439.2
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr7_+_150929550 | 0.17 |
ENST00000482173.1
ENST00000495645.1 ENST00000035307.2 |
CHPF2
|
chondroitin polymerizing factor 2 |
| chr10_-_22292675 | 0.17 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr10_+_75504105 | 0.17 |
ENST00000535742.1
ENST00000546025.1 ENST00000345254.4 ENST00000540668.1 ENST00000339365.2 ENST00000411652.2 |
SEC24C
|
SEC24 family member C |
| chr1_-_206945830 | 0.17 |
ENST00000423557.1
|
IL10
|
interleukin 10 |
| chr11_-_68039364 | 0.16 |
ENST00000533310.1
ENST00000304271.6 ENST00000527280.1 |
C11orf24
|
chromosome 11 open reading frame 24 |
| chr3_+_19988885 | 0.16 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr10_-_27444143 | 0.16 |
ENST00000477432.1
|
YME1L1
|
YME1-like 1 ATPase |
| chr12_-_50616122 | 0.16 |
ENST00000552823.1
ENST00000552909.1 |
LIMA1
|
LIM domain and actin binding 1 |
| chr20_+_57267669 | 0.16 |
ENST00000356091.6
|
NPEPL1
|
aminopeptidase-like 1 |
| chr6_+_26020672 | 0.16 |
ENST00000357647.3
|
HIST1H3A
|
histone cluster 1, H3a |
| chr17_-_48450534 | 0.16 |
ENST00000503633.1
ENST00000442592.3 ENST00000225969.4 |
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr19_+_797443 | 0.16 |
ENST00000394601.4
ENST00000589575.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr1_+_119911425 | 0.15 |
ENST00000361035.4
ENST00000325945.3 |
HAO2
|
hydroxyacid oxidase 2 (long chain) |
| chr1_+_11866270 | 0.15 |
ENST00000376497.3
ENST00000376487.3 ENST00000376496.3 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr5_-_39425068 | 0.15 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr12_+_4918342 | 0.15 |
ENST00000280684.3
ENST00000433855.1 |
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6 |
| chr3_+_132378741 | 0.15 |
ENST00000493720.2
|
UBA5
|
ubiquitin-like modifier activating enzyme 5 |
| chr19_-_41256207 | 0.15 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr11_-_65624415 | 0.15 |
ENST00000524553.1
ENST00000527344.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr1_+_228327923 | 0.15 |
ENST00000391865.3
|
GUK1
|
guanylate kinase 1 |
| chr3_-_4508925 | 0.15 |
ENST00000534863.1
ENST00000383843.5 ENST00000458465.2 ENST00000405420.2 ENST00000272902.5 |
SUMF1
|
sulfatase modifying factor 1 |
| chr20_+_18118703 | 0.15 |
ENST00000464792.1
|
CSRP2BP
|
CSRP2 binding protein |
| chr1_+_228327943 | 0.15 |
ENST00000366726.1
ENST00000312726.4 ENST00000366728.2 ENST00000453943.1 ENST00000366723.1 ENST00000366722.1 ENST00000435153.1 ENST00000366721.1 |
GUK1
|
guanylate kinase 1 |
| chr7_+_100271446 | 0.14 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr5_-_176730676 | 0.14 |
ENST00000393611.2
ENST00000303251.6 ENST00000303270.6 |
RAB24
|
RAB24, member RAS oncogene family |
| chr12_+_41086215 | 0.14 |
ENST00000547702.1
ENST00000551424.1 |
CNTN1
|
contactin 1 |
| chr3_-_57583052 | 0.14 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr8_-_125384927 | 0.14 |
ENST00000297632.6
|
TMEM65
|
transmembrane protein 65 |
| chr6_+_63921351 | 0.14 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr12_-_56320887 | 0.14 |
ENST00000398213.4
|
WIBG
|
within bgcn homolog (Drosophila) |
| chr4_+_25235597 | 0.14 |
ENST00000264864.6
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta |
| chr2_+_171785824 | 0.14 |
ENST00000452526.2
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr6_-_101329191 | 0.14 |
ENST00000324723.6
ENST00000369162.2 ENST00000522650.1 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chrX_-_7895479 | 0.14 |
ENST00000381042.4
|
PNPLA4
|
patatin-like phospholipase domain containing 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB3L1_CREB3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.4 | 3.6 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.4 | 1.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.3 | 1.5 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 1.7 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.3 | 0.8 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.3 | 1.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.2 | 1.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 1.3 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.2 | 1.3 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.2 | 1.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 0.7 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.6 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.5 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.4 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.3 | GO:1903519 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) positive regulation of apoptotic DNA fragmentation(GO:1902512) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 | 0.3 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.5 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.2 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.1 | 1.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.7 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 | 0.4 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 1.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.3 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.1 | 0.4 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 1.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.9 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.8 | GO:0007135 | meiosis II(GO:0007135) |
| 0.1 | 0.2 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.6 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.3 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.1 | 0.2 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.1 | 0.9 | GO:0000050 | urea cycle(GO:0000050) |
| 0.1 | 0.3 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 0.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.6 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.3 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.3 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.4 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.6 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.6 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.9 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.3 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.9 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.2 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.2 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.5 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.2 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 1.8 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.3 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.3 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.1 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:2000784 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 2.8 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.5 | GO:0048070 | regulation of developmental pigmentation(GO:0048070) |
| 0.0 | 1.4 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.2 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.5 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.2 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.7 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.0 | GO:0035993 | subthalamus development(GO:0021539) subthalamic nucleus development(GO:0021763) deltoid tuberosity development(GO:0035993) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.0 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.0 | GO:1903121 | central nervous system myelin maintenance(GO:0032286) regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:2000767 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.0 | GO:1903371 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.4 | 1.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.3 | 1.3 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 0.7 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 0.5 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 0.5 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 1.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.5 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.6 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 3.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 1.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.5 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.9 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.6 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 1.6 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.3 | 1.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.2 | 1.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 1.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 1.8 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 1.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.9 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.9 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.3 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.5 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.3 | GO:0008478 | pyridoxal kinase activity(GO:0008478) lithium ion binding(GO:0031403) |
| 0.1 | 0.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.4 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.2 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 1.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 4.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.0 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.2 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.3 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.4 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.6 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 1.8 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0098811 | RNA polymerase II transcription coactivator activity(GO:0001105) transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.0 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 2.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 2.7 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |