Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
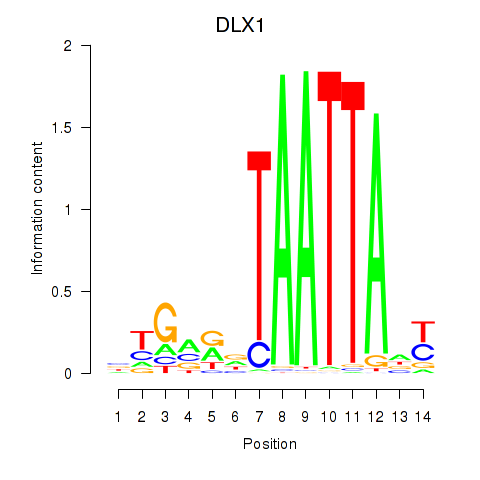
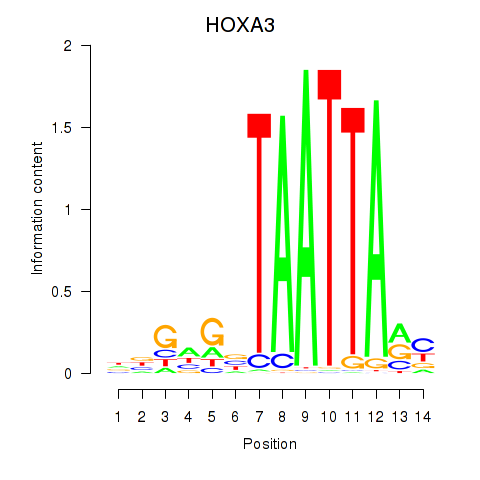
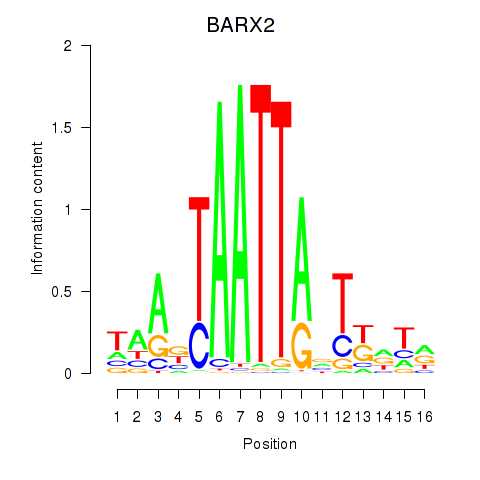
Results for DLX1_HOXA3_BARX2
Z-value: 1.25
Transcription factors associated with DLX1_HOXA3_BARX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX1
|
ENSG00000144355.10 | distal-less homeobox 1 |
|
HOXA3
|
ENSG00000105997.18 | homeobox A3 |
|
BARX2
|
ENSG00000043039.5 | BARX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BARX2 | hg19_v2_chr11_+_129245796_129245835 | -0.85 | 3.6e-09 | Click! |
| DLX1 | hg19_v2_chr2_+_172950227_172950264 | -0.60 | 4.4e-04 | Click! |
Activity profile of DLX1_HOXA3_BARX2 motif
Sorted Z-values of DLX1_HOXA3_BARX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_228736335 | 23.75 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr11_+_101918153 | 21.04 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr2_+_228736321 | 18.63 |
ENST00000309931.2
|
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr12_+_7013897 | 14.00 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr12_+_7014064 | 13.92 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr11_-_26593779 | 9.26 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr13_+_36050881 | 9.20 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr12_+_7014126 | 9.11 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr11_-_63376013 | 7.04 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr11_-_26593649 | 6.96 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr11_-_26593677 | 6.96 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr16_-_28634874 | 6.15 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr4_-_70518941 | 5.69 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr12_-_25348007 | 5.31 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr2_+_228735763 | 5.17 |
ENST00000373666.2
|
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr6_-_32908792 | 4.96 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr2_+_26624775 | 4.84 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr12_+_20963632 | 4.78 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr4_+_70861647 | 4.61 |
ENST00000246895.4
ENST00000381060.2 |
STATH
|
statherin |
| chr16_-_28621353 | 4.14 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr4_-_138453559 | 3.86 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr4_-_100356551 | 3.81 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr6_-_28411241 | 3.68 |
ENST00000289788.4
|
ZSCAN23
|
zinc finger and SCAN domain containing 23 |
| chr6_+_43612750 | 3.52 |
ENST00000372165.4
ENST00000372163.4 |
RSPH9
|
radial spoke head 9 homolog (Chlamydomonas) |
| chr12_+_20963647 | 3.34 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr10_-_13043697 | 3.20 |
ENST00000378825.3
|
CCDC3
|
coiled-coil domain containing 3 |
| chr19_+_56713670 | 3.18 |
ENST00000534327.1
|
ZSCAN5C
|
zinc finger and SCAN domain containing 5C |
| chr12_-_10282836 | 3.12 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr12_-_71533055 | 3.06 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chr19_+_50016610 | 3.05 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr20_+_43990576 | 3.01 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr11_+_71900703 | 2.98 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr4_+_41614909 | 2.92 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr11_+_71900572 | 2.91 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr4_-_25865159 | 2.87 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr19_+_50016411 | 2.78 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr19_-_41356347 | 2.67 |
ENST00000301141.5
|
CYP2A6
|
cytochrome P450, family 2, subfamily A, polypeptide 6 |
| chr5_-_41213607 | 2.54 |
ENST00000337836.5
ENST00000433294.1 |
C6
|
complement component 6 |
| chr3_+_138340049 | 2.53 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr16_+_82090028 | 2.51 |
ENST00000568090.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr16_+_53133070 | 2.51 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr5_-_137475071 | 2.42 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr16_-_28937027 | 2.39 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr5_-_160279207 | 2.37 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr10_-_27529486 | 2.35 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr10_-_118928543 | 2.28 |
ENST00000419373.2
|
RP11-501J20.2
|
RP11-501J20.2 |
| chr1_-_24741525 | 2.26 |
ENST00000374409.1
|
STPG1
|
sperm-tail PG-rich repeat containing 1 |
| chr15_-_55657428 | 2.25 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr7_+_99717230 | 2.22 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr4_-_100356291 | 2.21 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr11_+_27076764 | 2.19 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr6_+_26365443 | 2.19 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr5_-_20575959 | 2.18 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr14_-_106926724 | 2.18 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr9_-_117111222 | 2.17 |
ENST00000374079.4
|
AKNA
|
AT-hook transcription factor |
| chr14_-_25479811 | 2.17 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr9_-_5304432 | 2.08 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr12_-_15374343 | 2.00 |
ENST00000256953.2
ENST00000546331.1 |
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr12_-_10282742 | 2.00 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr14_-_54423529 | 2.00 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr7_+_116660246 | 1.98 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr5_+_140227048 | 1.97 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr12_-_10282681 | 1.96 |
ENST00000533022.1
|
CLEC7A
|
C-type lectin domain family 7, member A |
| chrX_-_13835147 | 1.94 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr6_+_131958436 | 1.94 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr3_+_152552685 | 1.89 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr12_-_10151773 | 1.89 |
ENST00000298527.6
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1, member B |
| chr1_+_180601139 | 1.88 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr4_-_87028478 | 1.83 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr11_-_128894053 | 1.83 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr8_-_10512569 | 1.79 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr1_+_47489240 | 1.78 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr1_-_150738261 | 1.78 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr19_+_49199209 | 1.76 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr16_-_28621312 | 1.75 |
ENST00000314752.7
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr6_-_87804815 | 1.75 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr9_-_5339873 | 1.71 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr21_-_32253874 | 1.66 |
ENST00000332378.4
|
KRTAP11-1
|
keratin associated protein 11-1 |
| chr12_-_15114658 | 1.65 |
ENST00000542276.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr10_+_114133773 | 1.65 |
ENST00000354655.4
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr14_-_78083112 | 1.62 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr3_-_121740969 | 1.61 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr3_-_100712352 | 1.61 |
ENST00000471714.1
ENST00000284322.5 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr6_-_52705641 | 1.60 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr4_+_41614720 | 1.59 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_+_160370344 | 1.59 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr5_+_140557371 | 1.55 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr3_-_191000172 | 1.48 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr4_-_66536196 | 1.48 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr14_-_90085458 | 1.47 |
ENST00000345097.4
ENST00000555855.1 ENST00000555353.1 |
FOXN3
|
forkhead box N3 |
| chr12_-_15114603 | 1.46 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr17_+_48823975 | 1.46 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr16_-_3350614 | 1.44 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr1_-_212965104 | 1.44 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr6_+_26402465 | 1.43 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr11_+_121447469 | 1.42 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr13_-_86373536 | 1.41 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr1_-_169396646 | 1.41 |
ENST00000367806.3
|
CCDC181
|
coiled-coil domain containing 181 |
| chr5_+_36608422 | 1.40 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr13_+_78315295 | 1.37 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr3_+_195447738 | 1.33 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr15_+_40697988 | 1.32 |
ENST00000487418.2
ENST00000479013.2 |
IVD
|
isovaleryl-CoA dehydrogenase |
| chr3_+_140396881 | 1.30 |
ENST00000286349.3
|
TRIM42
|
tripartite motif containing 42 |
| chr16_-_28621298 | 1.30 |
ENST00000566189.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr19_+_21579908 | 1.30 |
ENST00000596302.1
ENST00000392288.2 ENST00000594390.1 ENST00000355504.4 |
ZNF493
|
zinc finger protein 493 |
| chr16_-_28608364 | 1.30 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr7_-_130080977 | 1.30 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr10_+_94594351 | 1.29 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr22_+_18632666 | 1.29 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr15_-_37393406 | 1.29 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr8_+_105235572 | 1.27 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr6_-_32908765 | 1.27 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr4_+_159727272 | 1.26 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr7_-_64023441 | 1.26 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr5_-_42811986 | 1.26 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr4_-_70725856 | 1.26 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr10_+_695888 | 1.24 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr4_-_150736962 | 1.24 |
ENST00000502345.1
ENST00000510975.1 ENST00000511993.1 |
RP11-526A4.1
|
RP11-526A4.1 |
| chr3_-_112565703 | 1.24 |
ENST00000488794.1
|
CD200R1L
|
CD200 receptor 1-like |
| chr16_-_28608424 | 1.21 |
ENST00000335715.4
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr6_+_88106840 | 1.21 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164 |
| chr3_-_114790179 | 1.18 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr6_-_52710893 | 1.17 |
ENST00000284562.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr6_+_153552455 | 1.17 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr9_+_108463234 | 1.16 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr2_+_169757750 | 1.16 |
ENST00000375363.3
ENST00000429379.2 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase, catalytic, 2 |
| chr15_+_89631381 | 1.16 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr1_-_77685084 | 1.16 |
ENST00000370812.3
ENST00000359130.1 ENST00000445065.1 ENST00000370813.5 |
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr7_+_116654935 | 1.14 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr2_-_241497374 | 1.12 |
ENST00000373318.2
ENST00000406958.1 ENST00000391987.1 ENST00000373320.4 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chr4_+_69681710 | 1.12 |
ENST00000265403.7
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr6_-_110011704 | 1.12 |
ENST00000448084.2
|
AK9
|
adenylate kinase 9 |
| chrX_+_144908928 | 1.12 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr4_-_74486109 | 1.11 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr15_+_84115868 | 1.11 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr5_-_42812143 | 1.10 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr3_+_2933893 | 1.10 |
ENST00000397459.2
|
CNTN4
|
contactin 4 |
| chr11_+_27015628 | 1.10 |
ENST00000318627.2
|
FIBIN
|
fin bud initiation factor homolog (zebrafish) |
| chr7_+_142982023 | 1.08 |
ENST00000359333.3
ENST00000409244.1 ENST00000409541.1 ENST00000410004.1 |
TMEM139
|
transmembrane protein 139 |
| chr8_-_90996459 | 1.07 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr1_+_212965170 | 1.07 |
ENST00000532324.1
ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3
|
TatD DNase domain containing 3 |
| chr6_+_31895254 | 1.06 |
ENST00000299367.5
ENST00000442278.2 |
C2
|
complement component 2 |
| chr11_-_111649015 | 1.04 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr11_-_129062093 | 1.04 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr20_-_50418947 | 1.03 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr13_-_41593425 | 1.02 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr20_-_50418972 | 1.01 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr11_+_66276550 | 0.99 |
ENST00000419755.3
|
CTD-3074O7.11
|
Bardet-Biedl syndrome 1 protein |
| chr17_+_35851570 | 0.99 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr4_-_105416039 | 0.99 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr21_-_43346790 | 0.99 |
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chr1_-_190446759 | 0.99 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr1_+_236958554 | 0.98 |
ENST00000366577.5
ENST00000418145.2 |
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr4_+_77356248 | 0.98 |
ENST00000296043.6
|
SHROOM3
|
shroom family member 3 |
| chr1_-_149900122 | 0.96 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr7_-_92777606 | 0.96 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr4_+_113568207 | 0.96 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr10_-_28571015 | 0.96 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr6_-_109702885 | 0.95 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr5_-_96478466 | 0.95 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr5_-_150473127 | 0.95 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr10_-_113943447 | 0.94 |
ENST00000369425.1
ENST00000348367.4 ENST00000423155.1 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr6_+_26440700 | 0.94 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr3_+_108541545 | 0.93 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr15_+_49462434 | 0.92 |
ENST00000558145.1
ENST00000543495.1 ENST00000544523.1 ENST00000560138.1 |
GALK2
|
galactokinase 2 |
| chr10_+_7745232 | 0.92 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr1_+_101003687 | 0.91 |
ENST00000315033.4
|
GPR88
|
G protein-coupled receptor 88 |
| chr17_+_79953310 | 0.89 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr14_+_21387491 | 0.88 |
ENST00000258817.2
|
RP11-84C10.2
|
RP11-84C10.2 |
| chrX_+_130192318 | 0.86 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr16_+_15489603 | 0.84 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr14_-_61124977 | 0.83 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr17_+_1674982 | 0.80 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr3_+_186353756 | 0.80 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr14_+_22739823 | 0.80 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr14_-_92413353 | 0.79 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr12_-_15114191 | 0.78 |
ENST00000541380.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr7_-_99717463 | 0.78 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr19_-_22018966 | 0.77 |
ENST00000599906.1
ENST00000354959.4 |
ZNF43
|
zinc finger protein 43 |
| chr1_-_150669500 | 0.77 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr10_-_47151341 | 0.77 |
ENST00000422732.2
|
LINC00842
|
long intergenic non-protein coding RNA 842 |
| chr1_-_232651312 | 0.76 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr19_-_36606181 | 0.75 |
ENST00000221859.4
|
POLR2I
|
polymerase (RNA) II (DNA directed) polypeptide I, 14.5kDa |
| chr11_-_77791156 | 0.74 |
ENST00000281031.4
|
NDUFC2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa |
| chr2_+_234826016 | 0.74 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr11_+_57480046 | 0.73 |
ENST00000378312.4
ENST00000278422.4 |
TMX2
|
thioredoxin-related transmembrane protein 2 |
| chr14_+_22465771 | 0.73 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr4_-_66536057 | 0.72 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr1_-_92952433 | 0.72 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr15_-_72563585 | 0.72 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr12_-_15114492 | 0.72 |
ENST00000541546.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr4_+_146539415 | 0.71 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr4_-_74486347 | 0.71 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr9_+_2159850 | 0.71 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr15_+_49462397 | 0.71 |
ENST00000396509.2
|
GALK2
|
galactokinase 2 |
| chr2_-_213403565 | 0.71 |
ENST00000342788.4
ENST00000436443.1 |
ERBB4
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 |
| chr3_+_138340067 | 0.71 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr14_+_100485712 | 0.70 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr17_+_67498538 | 0.69 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr12_-_23737534 | 0.69 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX1_HOXA3_BARX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 2.0 | 6.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 2.0 | 5.9 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.7 | 16.6 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.7 | 7.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.7 | 2.7 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.7 | 2.0 | GO:0061149 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.7 | 4.6 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.6 | 5.8 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.6 | 1.9 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.6 | 2.5 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.5 | 1.6 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.5 | 1.6 | GO:0061341 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.5 | 1.4 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.4 | 2.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.4 | 1.7 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.4 | 0.8 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.4 | 1.9 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.4 | 1.1 | GO:0061566 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.3 | 50.5 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.3 | 2.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 1.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.3 | 4.6 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.3 | 0.9 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.3 | 5.8 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.3 | 3.5 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.3 | 1.3 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.3 | 20.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 0.7 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.2 | 0.9 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.2 | 0.7 | GO:2000366 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.2 | 1.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 0.7 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 5.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 1.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 0.9 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 1.9 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 0.8 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.2 | 2.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 2.4 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.2 | 1.4 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 2.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 1.9 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 0.6 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.2 | 0.5 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.2 | 1.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 2.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 0.7 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.2 | 1.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 1.3 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 7.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 0.5 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.1 | 0.7 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 1.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 2.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 2.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.7 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 1.8 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.4 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.1 | 0.3 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.4 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.1 | 0.4 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 | 0.2 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.8 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.1 | 2.8 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.5 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.1 | 1.0 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.9 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.9 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.4 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.7 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 2.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 2.8 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.1 | 0.2 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.1 | 0.7 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.9 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.3 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.8 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.4 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 1.0 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.4 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.1 | 0.1 | GO:1902908 | regulation of melanosome transport(GO:1902908) |
| 0.1 | 0.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.5 | GO:1904798 | regulation of core promoter binding(GO:1904796) positive regulation of core promoter binding(GO:1904798) |
| 0.1 | 0.4 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 1.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 1.8 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 1.9 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 0.3 | GO:1904685 | positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.1 | 1.6 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.2 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.5 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.1 | 0.2 | GO:1900111 | regulation of histone H3-K9 dimethylation(GO:1900109) positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.1 | 0.7 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.3 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.1 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 2.4 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.3 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 1.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 2.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.2 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 1.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.4 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 7.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.6 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.3 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.3 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.5 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.9 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.6 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) response to insulin-like growth factor stimulus(GO:1990418) |
| 0.0 | 1.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.3 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.2 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.7 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.3 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 2.3 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 1.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.0 | 0.2 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.0 | 0.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0035377 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 2.8 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 0.5 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 2.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.0 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 1.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.0 | 1.0 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.4 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.2 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.5 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.2 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 1.4 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 1.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.8 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.4 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.3 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.7 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.5 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.0 | 0.4 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.2 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 0.1 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.4 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.1 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 1.1 | GO:0090277 | positive regulation of peptide hormone secretion(GO:0090277) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 1.5 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.0 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0060187 | cell pole(GO:0060187) |
| 0.3 | 6.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 1.4 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.3 | 2.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 5.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 1.8 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.2 | 1.6 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 1.4 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 1.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 0.9 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 1.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 23.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 3.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 8.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.4 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 2.1 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 1.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 48.0 | GO:0005929 | cilium(GO:0005929) |
| 0.1 | 0.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.8 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 3.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 1.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 1.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 10.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 2.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.8 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 1.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 4.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 1.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.8 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.7 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 1.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 17.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 1.5 | 4.6 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 1.5 | 5.9 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 1.5 | 5.8 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 1.4 | 7.0 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 1.3 | 5.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.6 | 2.5 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.5 | 2.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.5 | 1.6 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.5 | 1.6 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.5 | 1.9 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.5 | 1.9 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.5 | 2.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.4 | 4.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 1.6 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.3 | 0.9 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 1.8 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 6.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.3 | 1.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 2.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 0.7 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.2 | 1.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 6.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 0.6 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.2 | 1.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 5.2 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.2 | 0.6 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.2 | 0.9 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.2 | 0.5 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 3.7 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 0.5 | GO:0036317 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.2 | 0.5 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 0.9 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 2.0 | GO:0070700 | co-receptor binding(GO:0039706) BMP receptor binding(GO:0070700) |
| 0.2 | 6.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.7 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 1.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 1.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 3.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.8 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 1.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.4 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 2.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.9 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.7 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.6 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.2 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.4 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 2.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 2.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.5 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 1.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 2.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 1.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 1.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 3.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 3.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.6 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 1.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 27.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 2.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 3.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 3.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 23.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.6 | 16.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.5 | 7.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 6.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 7.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 6.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 3.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 1.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.6 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.8 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.4 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 3.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |