Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
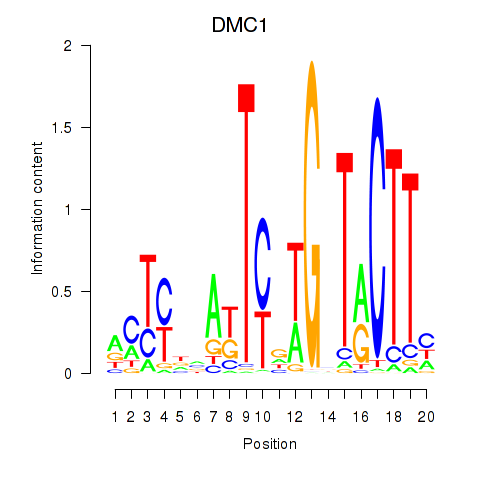
Results for DMC1
Z-value: 0.51
Transcription factors associated with DMC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DMC1
|
ENSG00000100206.5 | DNA meiotic recombinase 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DMC1 | hg19_v2_chr22_-_38966172_38966291 | -0.37 | 4.5e-02 | Click! |
Activity profile of DMC1 motif
Sorted Z-values of DMC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_94727048 | 3.85 |
ENST00000283357.5
|
FAM81B
|
family with sequence similarity 81, member B |
| chr7_+_6793740 | 2.54 |
ENST00000403107.1
ENST00000404077.1 ENST00000435395.1 ENST00000418406.1 |
RSPH10B2
|
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chr7_-_6010263 | 2.53 |
ENST00000455618.2
ENST00000405415.1 ENST00000404406.1 ENST00000542644.1 |
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr4_+_15471489 | 2.00 |
ENST00000424120.1
ENST00000413206.1 ENST00000438599.2 ENST00000511544.1 ENST00000512702.1 ENST00000507954.1 ENST00000515124.1 ENST00000503292.1 ENST00000503658.1 |
CC2D2A
|
coiled-coil and C2 domain containing 2A |
| chr21_-_46348694 | 1.75 |
ENST00000355153.4
ENST00000397850.2 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr2_+_120187465 | 1.22 |
ENST00000409826.1
ENST00000417645.1 |
TMEM37
|
transmembrane protein 37 |
| chr10_-_62332357 | 1.02 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr11_+_101785727 | 1.00 |
ENST00000263468.8
|
KIAA1377
|
KIAA1377 |
| chr10_+_74870206 | 0.90 |
ENST00000357321.4
ENST00000349051.5 |
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr10_+_74870253 | 0.81 |
ENST00000544879.1
ENST00000537969.1 ENST00000372997.3 |
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chrX_+_105855160 | 0.74 |
ENST00000372544.2
ENST00000372548.4 |
CXorf57
|
chromosome X open reading frame 57 |
| chr1_+_166958346 | 0.73 |
ENST00000367872.4
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr11_+_71238313 | 0.72 |
ENST00000398536.4
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr12_-_9268707 | 0.67 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr1_+_166958497 | 0.63 |
ENST00000367870.2
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr2_+_233497931 | 0.58 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr20_-_21378666 | 0.47 |
ENST00000351817.4
|
NKX2-4
|
NK2 homeobox 4 |
| chr20_+_11008408 | 0.47 |
ENST00000378252.1
|
C20orf187
|
chromosome 20 open reading frame 187 |
| chr14_+_74318611 | 0.45 |
ENST00000555976.1
ENST00000267568.4 |
PTGR2
|
prostaglandin reductase 2 |
| chr14_+_74318513 | 0.45 |
ENST00000555228.1
ENST00000555661.1 |
PTGR2
|
prostaglandin reductase 2 |
| chr2_-_18741882 | 0.44 |
ENST00000381249.3
|
RDH14
|
retinol dehydrogenase 14 (all-trans/9-cis/11-cis) |
| chr14_-_21492113 | 0.41 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr14_-_21492251 | 0.37 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr3_-_108672742 | 0.36 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr3_-_108672609 | 0.35 |
ENST00000393963.3
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chrX_+_65382433 | 0.34 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr14_-_21491477 | 0.34 |
ENST00000298684.5
ENST00000557169.1 ENST00000553563.1 |
NDRG2
|
NDRG family member 2 |
| chr17_+_77018896 | 0.34 |
ENST00000578229.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_+_77019030 | 0.29 |
ENST00000580454.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr4_+_106473768 | 0.29 |
ENST00000265154.2
ENST00000420470.2 |
ARHGEF38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr5_-_135231516 | 0.27 |
ENST00000274520.1
|
IL9
|
interleukin 9 |
| chr1_-_182642017 | 0.22 |
ENST00000367557.4
ENST00000258302.4 |
RGS8
|
regulator of G-protein signaling 8 |
| chr10_-_74856608 | 0.21 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr16_-_30122717 | 0.21 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr9_-_34729457 | 0.20 |
ENST00000378788.3
|
FAM205A
|
family with sequence similarity 205, member A |
| chr6_+_80129989 | 0.19 |
ENST00000429444.1
|
RP1-232L24.3
|
RP1-232L24.3 |
| chrX_-_134305322 | 0.18 |
ENST00000276241.6
ENST00000344129.2 |
CXorf48
|
cancer/testis antigen 55 |
| chrX_+_65382381 | 0.18 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr1_-_47131521 | 0.18 |
ENST00000542495.1
ENST00000532925.1 |
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr6_-_31670723 | 0.17 |
ENST00000440843.2
ENST00000375842.4 |
ABHD16A
|
abhydrolase domain containing 16A |
| chr15_-_49103235 | 0.17 |
ENST00000380950.2
|
CEP152
|
centrosomal protein 152kDa |
| chr10_-_98031265 | 0.16 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr10_-_98031310 | 0.14 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr11_-_73720122 | 0.14 |
ENST00000426995.2
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr10_+_17851362 | 0.13 |
ENST00000331429.2
ENST00000457317.1 |
MRC1L1
|
cDNA FLJ56855, highly similar to Macrophage mannose receptor 1 |
| chr11_+_71846748 | 0.12 |
ENST00000445078.2
|
FOLR3
|
folate receptor 3 (gamma) |
| chr17_-_60142609 | 0.11 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr1_+_18958008 | 0.11 |
ENST00000420770.2
ENST00000400661.3 |
PAX7
|
paired box 7 |
| chr12_-_71031220 | 0.11 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr6_-_62996066 | 0.10 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr2_-_163008903 | 0.10 |
ENST00000418842.2
ENST00000375497.3 |
GCG
|
glucagon |
| chr11_+_71846764 | 0.10 |
ENST00000456237.1
ENST00000442948.2 ENST00000546166.1 |
FOLR3
|
folate receptor 3 (gamma) |
| chr12_-_53901266 | 0.09 |
ENST00000609999.1
ENST00000267017.3 |
NPFF
|
neuropeptide FF-amide peptide precursor |
| chr10_+_62538248 | 0.07 |
ENST00000448257.2
|
CDK1
|
cyclin-dependent kinase 1 |
| chr17_+_4846101 | 0.07 |
ENST00000576965.1
|
RNF167
|
ring finger protein 167 |
| chr19_+_44617511 | 0.06 |
ENST00000262894.6
ENST00000588926.1 ENST00000592780.1 |
ZNF225
|
zinc finger protein 225 |
| chr2_+_219246746 | 0.06 |
ENST00000233202.6
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr5_-_157603408 | 0.05 |
ENST00000522769.1
|
CTC-436K13.5
|
CTC-436K13.5 |
| chr12_-_71031185 | 0.04 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr10_+_62538089 | 0.04 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr1_-_11863571 | 0.04 |
ENST00000376583.3
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr19_-_3600549 | 0.04 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chr7_+_47694842 | 0.03 |
ENST00000408988.2
|
C7orf65
|
chromosome 7 open reading frame 65 |
| chr21_-_35281399 | 0.03 |
ENST00000418933.1
|
ATP5O
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr19_-_42947121 | 0.02 |
ENST00000601181.1
|
CXCL17
|
chemokine (C-X-C motif) ligand 17 |
| chr3_+_9944492 | 0.02 |
ENST00000383814.3
ENST00000454190.2 ENST00000454992.1 |
IL17RE
|
interleukin 17 receptor E |
| chr8_-_101734907 | 0.02 |
ENST00000318607.5
ENST00000521865.1 ENST00000520804.1 ENST00000522720.1 ENST00000521067.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr20_+_2796948 | 0.02 |
ENST00000361033.1
ENST00000380585.1 |
TMEM239
|
transmembrane protein 239 |
| chr5_+_55149356 | 0.02 |
ENST00000490985.1
ENST00000354961.4 |
IL31RA
|
interleukin 31 receptor A |
| chr3_+_9944303 | 0.01 |
ENST00000421412.1
ENST00000295980.3 |
IL17RE
|
interleukin 17 receptor E |
| chr10_-_135187193 | 0.00 |
ENST00000368547.3
|
ECHS1
|
enoyl CoA hydratase, short chain, 1, mitochondrial |
| chr16_+_2255710 | 0.00 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of DMC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.3 | 1.0 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.6 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 1.1 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 1.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.4 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.7 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 1.7 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.1 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.2 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.0 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.0 | GO:0055073 | cadmium ion homeostasis(GO:0055073) divalent metal ion export(GO:0070839) |
| 0.0 | 0.6 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0030849 | autosome(GO:0030849) |
| 0.2 | 1.4 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 2.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 1.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 1.4 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 0.9 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 0.7 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 1.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.0 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |