Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for DPRX
Z-value: 0.63
Transcription factors associated with DPRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DPRX
|
ENSG00000204595.1 | divergent-paired related homeobox |
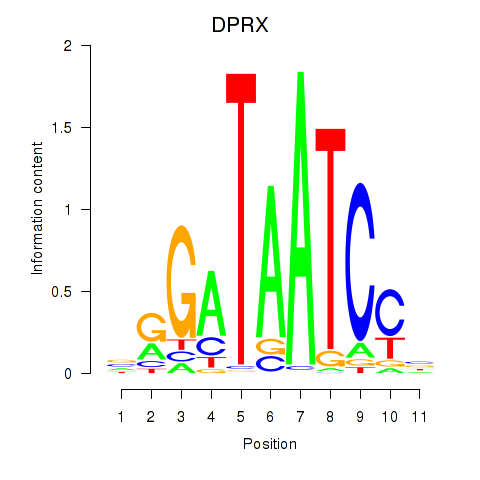
Activity profile of DPRX motif
Sorted Z-values of DPRX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_123241908 | 1.66 |
ENST00000434204.1
ENST00000437535.1 ENST00000451215.1 |
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr1_-_150208320 | 1.27 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr3_-_112564797 | 0.79 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr11_-_321340 | 0.73 |
ENST00000526811.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr9_-_13279406 | 0.71 |
ENST00000546205.1
|
MPDZ
|
multiple PDZ domain protein |
| chr9_-_13279563 | 0.71 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chrX_+_53449805 | 0.70 |
ENST00000414955.2
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chrX_+_53449887 | 0.70 |
ENST00000375327.3
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chr4_+_159131630 | 0.64 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chr1_-_46089639 | 0.64 |
ENST00000445048.2
|
CCDC17
|
coiled-coil domain containing 17 |
| chr4_+_36283213 | 0.61 |
ENST00000357504.3
|
DTHD1
|
death domain containing 1 |
| chr16_-_28415162 | 0.58 |
ENST00000398944.3
ENST00000398943.3 ENST00000380876.4 |
EIF3CL
|
eukaryotic translation initiation factor 3, subunit C-like |
| chr16_+_28722684 | 0.54 |
ENST00000331666.6
ENST00000395587.1 ENST00000569690.1 ENST00000564243.1 |
EIF3C
|
eukaryotic translation initiation factor 3, subunit C |
| chr16_+_28722809 | 0.52 |
ENST00000566866.1
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C |
| chr8_-_79717750 | 0.52 |
ENST00000263851.4
ENST00000379113.2 |
IL7
|
interleukin 7 |
| chr11_-_111944895 | 0.52 |
ENST00000431456.1
ENST00000280350.4 ENST00000530641.1 |
PIH1D2
|
PIH1 domain containing 2 |
| chr1_+_179561011 | 0.50 |
ENST00000294848.8
ENST00000444136.1 |
TDRD5
|
tudor domain containing 5 |
| chr3_-_167371740 | 0.49 |
ENST00000466760.1
ENST00000479765.1 |
WDR49
|
WD repeat domain 49 |
| chr11_+_111385497 | 0.46 |
ENST00000375618.4
ENST00000529167.1 ENST00000332814.6 |
C11orf88
|
chromosome 11 open reading frame 88 |
| chr11_-_321050 | 0.46 |
ENST00000399808.4
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr2_+_33701286 | 0.45 |
ENST00000403687.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr4_+_169418255 | 0.42 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr17_+_3118915 | 0.42 |
ENST00000304094.1
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1 |
| chr14_-_22005343 | 0.42 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr11_-_111944704 | 0.41 |
ENST00000532211.1
|
PIH1D2
|
PIH1 domain containing 2 |
| chr11_-_124190184 | 0.41 |
ENST00000357438.2
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2 |
| chr6_-_37225391 | 0.41 |
ENST00000356757.2
|
TMEM217
|
transmembrane protein 217 |
| chr16_-_50913188 | 0.40 |
ENST00000569986.1
|
CTD-2034I21.1
|
CTD-2034I21.1 |
| chr16_-_30022735 | 0.40 |
ENST00000564944.1
|
DOC2A
|
double C2-like domains, alpha |
| chr6_-_37225367 | 0.40 |
ENST00000336655.2
|
TMEM217
|
transmembrane protein 217 |
| chr7_-_99869799 | 0.40 |
ENST00000436886.2
|
GATS
|
GATS, stromal antigen 3 opposite strand |
| chr3_+_45984947 | 0.39 |
ENST00000304552.4
|
CXCR6
|
chemokine (C-X-C motif) receptor 6 |
| chr8_+_8559406 | 0.39 |
ENST00000519106.1
|
CLDN23
|
claudin 23 |
| chr4_-_123542224 | 0.39 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr18_+_56530136 | 0.38 |
ENST00000591083.1
|
ZNF532
|
zinc finger protein 532 |
| chr7_+_28452130 | 0.36 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr6_-_32784687 | 0.36 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr1_-_55089191 | 0.35 |
ENST00000302250.2
ENST00000371304.2 |
FAM151A
|
family with sequence similarity 151, member A |
| chr16_-_20556492 | 0.34 |
ENST00000568098.1
|
ACSM2B
|
acyl-CoA synthetase medium-chain family member 2B |
| chr7_-_94285511 | 0.34 |
ENST00000265735.7
|
SGCE
|
sarcoglycan, epsilon |
| chr1_-_236046872 | 0.34 |
ENST00000536965.1
|
LYST
|
lysosomal trafficking regulator |
| chr14_-_22005062 | 0.33 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr3_+_49058444 | 0.32 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr2_+_183943464 | 0.30 |
ENST00000354221.4
|
DUSP19
|
dual specificity phosphatase 19 |
| chr3_+_158991025 | 0.29 |
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr1_-_13452656 | 0.29 |
ENST00000376132.3
|
PRAMEF13
|
PRAME family member 13 |
| chr7_+_150264365 | 0.29 |
ENST00000255945.2
ENST00000461940.1 |
GIMAP4
|
GTPase, IMAP family member 4 |
| chr3_+_148447887 | 0.28 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr17_-_1928621 | 0.27 |
ENST00000331238.6
|
RTN4RL1
|
reticulon 4 receptor-like 1 |
| chr11_+_12399071 | 0.27 |
ENST00000539723.1
ENST00000550549.1 |
PARVA
|
parvin, alpha |
| chr14_+_85996471 | 0.26 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chrX_+_15525426 | 0.26 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr3_+_97483366 | 0.26 |
ENST00000463745.1
ENST00000462412.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr6_-_32731243 | 0.26 |
ENST00000427449.1
ENST00000411527.1 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr12_-_103310987 | 0.26 |
ENST00000307000.2
|
PAH
|
phenylalanine hydroxylase |
| chr15_+_34261089 | 0.25 |
ENST00000383263.5
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr2_-_119605253 | 0.25 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr4_-_174451370 | 0.25 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr19_-_22806764 | 0.25 |
ENST00000598042.1
|
AC011516.2
|
AC011516.2 |
| chr1_+_236557569 | 0.24 |
ENST00000334232.4
|
EDARADD
|
EDAR-associated death domain |
| chr3_-_49058479 | 0.24 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr10_+_104178946 | 0.24 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr14_+_70918874 | 0.24 |
ENST00000603540.1
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr2_+_33661382 | 0.24 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr19_-_50370799 | 0.24 |
ENST00000600910.1
ENST00000322344.3 ENST00000600573.1 |
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr1_-_153521597 | 0.24 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr11_-_2323290 | 0.23 |
ENST00000381153.3
|
C11orf21
|
chromosome 11 open reading frame 21 |
| chr9_+_17134980 | 0.23 |
ENST00000380647.3
|
CNTLN
|
centlein, centrosomal protein |
| chr8_+_54764346 | 0.23 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr6_-_22297730 | 0.22 |
ENST00000306482.1
|
PRL
|
prolactin |
| chr7_-_135412925 | 0.22 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr3_-_158390282 | 0.22 |
ENST00000264265.3
|
LXN
|
latexin |
| chr8_-_72268889 | 0.21 |
ENST00000388742.4
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chrX_+_53449839 | 0.21 |
ENST00000457095.1
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chr8_-_72268968 | 0.21 |
ENST00000388740.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr16_-_67427389 | 0.21 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr6_-_132967142 | 0.21 |
ENST00000275216.1
|
TAAR1
|
trace amine associated receptor 1 |
| chr3_-_18466787 | 0.21 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr3_-_45883558 | 0.21 |
ENST00000445698.1
ENST00000296135.6 |
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr5_-_180669236 | 0.21 |
ENST00000507756.1
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr19_-_57967854 | 0.20 |
ENST00000321039.3
|
VN1R1
|
vomeronasal 1 receptor 1 |
| chr2_-_26700900 | 0.20 |
ENST00000338581.6
ENST00000339598.3 ENST00000402415.3 |
OTOF
|
otoferlin |
| chr17_-_19771216 | 0.20 |
ENST00000395544.4
|
ULK2
|
unc-51 like autophagy activating kinase 2 |
| chr6_-_170599561 | 0.20 |
ENST00000366756.3
|
DLL1
|
delta-like 1 (Drosophila) |
| chr2_-_235405168 | 0.19 |
ENST00000339728.3
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr9_+_37667978 | 0.19 |
ENST00000539465.1
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chrX_+_46771711 | 0.19 |
ENST00000424392.1
ENST00000397189.1 |
PHF16
|
jade family PHD finger 3 |
| chr11_-_10715287 | 0.18 |
ENST00000423302.2
|
MRVI1
|
murine retrovirus integration site 1 homolog |
| chr6_+_46761118 | 0.18 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr17_-_19771242 | 0.18 |
ENST00000361658.2
|
ULK2
|
unc-51 like autophagy activating kinase 2 |
| chr12_+_130554803 | 0.18 |
ENST00000535487.1
|
RP11-474D1.2
|
RP11-474D1.2 |
| chr13_+_52436111 | 0.18 |
ENST00000242819.4
|
CCDC70
|
coiled-coil domain containing 70 |
| chr1_-_170043709 | 0.18 |
ENST00000367767.1
ENST00000361580.2 ENST00000538366.1 |
KIFAP3
|
kinesin-associated protein 3 |
| chr1_+_6615241 | 0.18 |
ENST00000333172.6
ENST00000328191.4 ENST00000351136.3 |
TAS1R1
|
taste receptor, type 1, member 1 |
| chr1_-_153521714 | 0.18 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr15_+_22382382 | 0.17 |
ENST00000328795.4
|
OR4N4
|
olfactory receptor, family 4, subfamily N, member 4 |
| chr1_-_154458520 | 0.17 |
ENST00000486773.1
|
SHE
|
Src homology 2 domain containing E |
| chr14_+_85996507 | 0.17 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr13_+_28712614 | 0.16 |
ENST00000380958.3
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr1_+_63989004 | 0.16 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr7_-_75401513 | 0.16 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr11_+_61248583 | 0.16 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr11_-_30608413 | 0.16 |
ENST00000528686.1
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr14_+_22670455 | 0.16 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr17_+_7482785 | 0.15 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr12_+_113344582 | 0.15 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr12_+_86268065 | 0.15 |
ENST00000551529.1
ENST00000256010.6 |
NTS
|
neurotensin |
| chr3_+_141105705 | 0.15 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr3_+_111393501 | 0.15 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr8_-_53626974 | 0.15 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chr11_+_64948665 | 0.15 |
ENST00000533820.1
|
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr14_+_62585332 | 0.15 |
ENST00000554895.1
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
| chrX_+_100333709 | 0.15 |
ENST00000372930.4
|
TMEM35
|
transmembrane protein 35 |
| chr21_-_28217721 | 0.15 |
ENST00000284984.3
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr22_-_26908343 | 0.15 |
ENST00000407431.1
ENST00000455080.1 ENST00000418876.1 ENST00000440258.1 ENST00000407148.1 ENST00000420242.1 ENST00000405938.1 ENST00000407690.1 |
TFIP11
|
tuftelin interacting protein 11 |
| chr8_-_101157680 | 0.14 |
ENST00000428847.2
|
FBXO43
|
F-box protein 43 |
| chr2_+_69201705 | 0.14 |
ENST00000377938.2
|
GKN1
|
gastrokine 1 |
| chr5_+_35856951 | 0.14 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr10_+_70661014 | 0.14 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr8_-_95274536 | 0.14 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr1_+_210501589 | 0.14 |
ENST00000413764.2
ENST00000541565.1 |
HHAT
|
hedgehog acyltransferase |
| chr1_-_94586651 | 0.14 |
ENST00000535735.1
ENST00000370225.3 |
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr2_-_49381572 | 0.14 |
ENST00000454032.1
ENST00000304421.4 |
FSHR
|
follicle stimulating hormone receptor |
| chr12_-_71182695 | 0.14 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr6_+_28317685 | 0.14 |
ENST00000252211.2
ENST00000341464.5 ENST00000377255.3 |
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3 |
| chr2_-_49381646 | 0.14 |
ENST00000346173.3
ENST00000406846.2 |
FSHR
|
follicle stimulating hormone receptor |
| chr5_+_138678131 | 0.14 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr11_-_77123065 | 0.13 |
ENST00000530617.1
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr11_+_64949343 | 0.13 |
ENST00000279247.6
ENST00000532285.1 ENST00000534373.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr20_+_22034809 | 0.13 |
ENST00000449427.1
|
RP11-125P18.1
|
RP11-125P18.1 |
| chr17_+_8191815 | 0.13 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr19_+_2476116 | 0.13 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr13_-_44980021 | 0.13 |
ENST00000432701.2
ENST00000607312.1 |
LINC01071
|
long intergenic non-protein coding RNA 1071 |
| chr2_+_128175997 | 0.13 |
ENST00000234071.3
ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr3_-_124653579 | 0.12 |
ENST00000478191.1
ENST00000311075.3 |
MUC13
|
mucin 13, cell surface associated |
| chr15_+_81475047 | 0.12 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chr12_-_56321397 | 0.12 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr18_-_40857493 | 0.12 |
ENST00000255224.3
|
SYT4
|
synaptotagmin IV |
| chr9_+_109625378 | 0.12 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr2_+_74685413 | 0.12 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
| chr18_-_3845293 | 0.12 |
ENST00000400145.2
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr19_-_7040190 | 0.12 |
ENST00000381394.4
|
MBD3L4
|
methyl-CpG binding domain protein 3-like 4 |
| chr8_-_10987745 | 0.12 |
ENST00000400102.3
|
AF131215.5
|
AF131215.5 |
| chrX_+_77166172 | 0.11 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr4_-_153700864 | 0.11 |
ENST00000304337.2
|
TIGD4
|
tigger transposable element derived 4 |
| chr2_+_234216454 | 0.11 |
ENST00000447536.1
ENST00000409110.1 |
SAG
|
S-antigen; retina and pineal gland (arrestin) |
| chr5_-_134871639 | 0.11 |
ENST00000314744.4
|
NEUROG1
|
neurogenin 1 |
| chr1_+_20396649 | 0.11 |
ENST00000375108.3
|
PLA2G5
|
phospholipase A2, group V |
| chr20_+_8112824 | 0.11 |
ENST00000378641.3
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr17_-_7080227 | 0.11 |
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr17_+_5323492 | 0.11 |
ENST00000405578.4
ENST00000574003.1 |
RPAIN
|
RPA interacting protein |
| chr6_-_31080336 | 0.11 |
ENST00000259870.3
|
C6orf15
|
chromosome 6 open reading frame 15 |
| chr5_+_138677515 | 0.11 |
ENST00000265192.4
ENST00000511706.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr8_+_1772132 | 0.11 |
ENST00000349830.3
ENST00000520359.1 ENST00000518288.1 ENST00000398560.1 |
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr16_+_30386098 | 0.11 |
ENST00000322861.7
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr1_+_16767167 | 0.11 |
ENST00000337132.5
|
NECAP2
|
NECAP endocytosis associated 2 |
| chr19_-_7058651 | 0.11 |
ENST00000333843.4
|
MBD3L3
|
methyl-CpG binding domain protein 3-like 3 |
| chr1_+_16767195 | 0.11 |
ENST00000504551.2
ENST00000457722.2 ENST00000406746.1 ENST00000443980.2 |
NECAP2
|
NECAP endocytosis associated 2 |
| chr8_+_145215928 | 0.10 |
ENST00000528919.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr19_+_7049332 | 0.10 |
ENST00000381393.3
|
MBD3L2
|
methyl-CpG binding domain protein 3-like 2 |
| chr2_-_190044480 | 0.10 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr3_-_18466026 | 0.10 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr7_-_97881429 | 0.10 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr14_+_61995722 | 0.10 |
ENST00000556347.1
|
RP11-47I22.4
|
RP11-47I22.4 |
| chr11_-_118436707 | 0.10 |
ENST00000264020.2
ENST00000264021.3 |
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr5_-_59064458 | 0.10 |
ENST00000502575.1
ENST00000507116.1 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr19_+_7030589 | 0.10 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3-like 5 |
| chrX_-_138724994 | 0.10 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr7_+_135242652 | 0.09 |
ENST00000285968.6
ENST00000440390.2 |
NUP205
|
nucleoporin 205kDa |
| chr6_-_10115007 | 0.09 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr4_-_87374283 | 0.09 |
ENST00000361569.2
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr12_+_122516626 | 0.09 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chr4_+_186125391 | 0.09 |
ENST00000504273.1
|
SNX25
|
sorting nexin 25 |
| chr1_+_202317815 | 0.09 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr14_-_94789663 | 0.09 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr1_+_120049826 | 0.09 |
ENST00000369413.3
ENST00000235547.6 ENST00000528909.1 |
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr9_-_112083229 | 0.09 |
ENST00000374566.3
ENST00000374557.4 |
EPB41L4B
|
erythrocyte membrane protein band 4.1 like 4B |
| chr14_+_22265444 | 0.09 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr4_+_170541835 | 0.09 |
ENST00000504131.2
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr7_+_94536898 | 0.08 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr11_-_104916034 | 0.08 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr11_-_104972158 | 0.08 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr11_-_118436606 | 0.08 |
ENST00000530872.1
|
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr1_+_207262578 | 0.08 |
ENST00000243611.5
ENST00000367076.3 |
C4BPB
|
complement component 4 binding protein, beta |
| chr20_+_123010 | 0.08 |
ENST00000382398.3
|
DEFB126
|
defensin, beta 126 |
| chr16_+_30406721 | 0.08 |
ENST00000320159.2
|
ZNF48
|
zinc finger protein 48 |
| chr7_-_155601766 | 0.07 |
ENST00000430104.1
|
SHH
|
sonic hedgehog |
| chr18_-_47376197 | 0.07 |
ENST00000592688.1
|
MYO5B
|
myosin VB |
| chr14_-_69446034 | 0.07 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr17_+_7531281 | 0.07 |
ENST00000575729.1
ENST00000340624.5 |
SHBG
|
sex hormone-binding globulin |
| chr14_+_75469606 | 0.07 |
ENST00000266126.5
|
EIF2B2
|
eukaryotic translation initiation factor 2B, subunit 2 beta, 39kDa |
| chr2_+_145425534 | 0.07 |
ENST00000432608.1
ENST00000597655.1 ENST00000598659.1 ENST00000600679.1 ENST00000601277.1 ENST00000451027.1 ENST00000445791.1 ENST00000596540.1 ENST00000596230.1 ENST00000594471.1 ENST00000598248.1 ENST00000597469.1 ENST00000431734.1 ENST00000595686.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr12_+_133563026 | 0.07 |
ENST00000540238.1
ENST00000328654.5 ENST00000544181.1 |
ZNF26
|
zinc finger protein 26 |
| chr1_+_207262627 | 0.07 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein, beta |
| chr2_+_27799389 | 0.07 |
ENST00000408964.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chr1_-_68516393 | 0.07 |
ENST00000395201.1
|
DIRAS3
|
DIRAS family, GTP-binding RAS-like 3 |
| chr10_+_90346519 | 0.07 |
ENST00000371939.3
|
LIPJ
|
lipase, family member J |
| chrX_-_70474499 | 0.07 |
ENST00000353904.2
|
ZMYM3
|
zinc finger, MYM-type 3 |
| chr1_+_19638788 | 0.07 |
ENST00000375155.3
ENST00000375153.3 ENST00000400548.2 |
PQLC2
|
PQ loop repeat containing 2 |
| chr1_+_207262170 | 0.07 |
ENST00000367078.3
|
C4BPB
|
complement component 4 binding protein, beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of DPRX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 1.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 0.3 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.2 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.3 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.1 | 0.3 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.2 | GO:0048633 | negative regulation of auditory receptor cell differentiation(GO:0045608) positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.4 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.2 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 1.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.5 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.4 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.2 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 1.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:2001033 | negative regulation of double-strand break repair via nonhomologous end joining(GO:2001033) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.3 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.3 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.4 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.0 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.0 | GO:1903179 | positive regulation of Wnt protein secretion(GO:0061357) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.6 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.0 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.1 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.5 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.1 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.0 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.4 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 1.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.5 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.2 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.3 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.1 | 0.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.3 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.2 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.6 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.4 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0016295 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 1.4 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.0 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |