Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
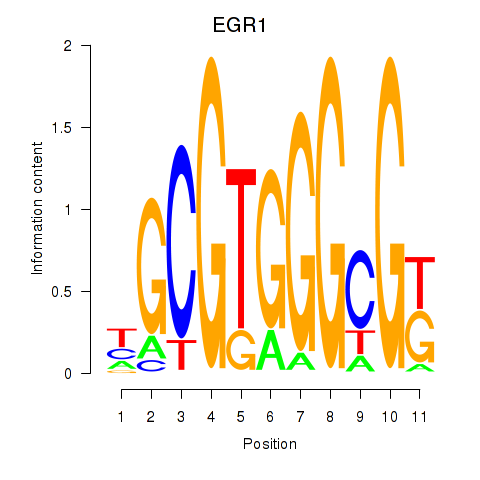
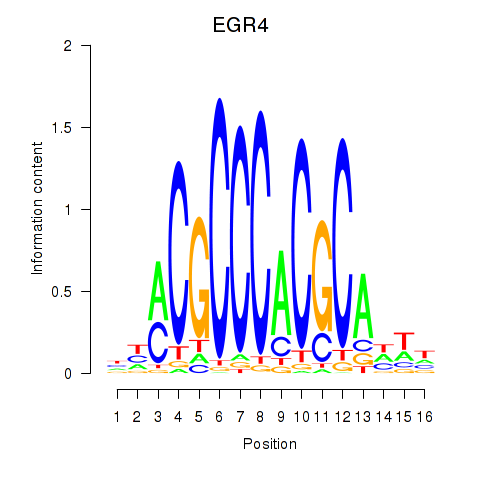
Results for EGR1_EGR4
Z-value: 0.98
Transcription factors associated with EGR1_EGR4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EGR1
|
ENSG00000120738.7 | early growth response 1 |
|
EGR4
|
ENSG00000135625.6 | early growth response 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EGR1 | hg19_v2_chr5_+_137801160_137801179 | 0.69 | 2.2e-05 | Click! |
| EGR4 | hg19_v2_chr2_-_73520667_73520833 | -0.09 | 6.4e-01 | Click! |
Activity profile of EGR1_EGR4 motif
Sorted Z-values of EGR1_EGR4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_57634475 | 9.77 |
ENST00000393825.1
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr6_-_4135825 | 6.82 |
ENST00000380118.3
ENST00000413766.2 ENST00000361538.2 |
ECI2
|
enoyl-CoA delta isomerase 2 |
| chr19_-_51472031 | 6.08 |
ENST00000391808.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr1_-_9189229 | 5.96 |
ENST00000377411.4
|
GPR157
|
G protein-coupled receptor 157 |
| chr19_-_51472222 | 5.28 |
ENST00000376851.3
|
KLK6
|
kallikrein-related peptidase 6 |
| chr15_-_80263506 | 4.31 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr15_+_39873268 | 4.20 |
ENST00000397591.2
ENST00000260356.5 |
THBS1
|
thrombospondin 1 |
| chr6_-_4135693 | 4.05 |
ENST00000495548.1
ENST00000380125.2 ENST00000465828.1 |
ECI2
|
enoyl-CoA delta isomerase 2 |
| chr1_+_65886244 | 3.92 |
ENST00000344610.8
|
LEPR
|
leptin receptor |
| chr2_-_27718052 | 3.82 |
ENST00000264703.3
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr6_+_86159821 | 3.80 |
ENST00000369651.3
|
NT5E
|
5'-nucleotidase, ecto (CD73) |
| chr6_+_151646800 | 3.78 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr1_+_23695680 | 3.58 |
ENST00000454117.1
ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213
|
chromosome 1 open reading frame 213 |
| chr11_-_2160180 | 3.45 |
ENST00000381406.4
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr15_-_90039805 | 3.41 |
ENST00000544600.1
ENST00000268122.4 |
RHCG
|
Rh family, C glycoprotein |
| chr15_+_74833518 | 3.33 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr1_-_94312706 | 3.30 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr10_-_129924611 | 3.28 |
ENST00000368654.3
|
MKI67
|
marker of proliferation Ki-67 |
| chr6_+_86159765 | 3.27 |
ENST00000369646.3
ENST00000257770.3 |
NT5E
|
5'-nucleotidase, ecto (CD73) |
| chr1_+_65886326 | 3.27 |
ENST00000371059.3
ENST00000371060.3 ENST00000349533.6 ENST00000406510.3 |
LEPR
|
leptin receptor |
| chr6_+_43739697 | 3.23 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr12_-_76425368 | 2.95 |
ENST00000602540.1
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1 |
| chr10_-_43762329 | 2.83 |
ENST00000395810.1
|
RASGEF1A
|
RasGEF domain family, member 1A |
| chr13_+_110959598 | 2.68 |
ENST00000360467.5
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr11_-_2160611 | 2.65 |
ENST00000416167.2
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr6_+_37137939 | 2.50 |
ENST00000373509.5
|
PIM1
|
pim-1 oncogene |
| chr16_+_58497567 | 2.47 |
ENST00000258187.5
|
NDRG4
|
NDRG family member 4 |
| chr5_-_176924562 | 2.43 |
ENST00000359895.2
ENST00000355572.2 ENST00000355841.2 ENST00000393551.1 ENST00000505074.1 ENST00000356618.4 ENST00000393546.4 |
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr19_+_45843994 | 2.37 |
ENST00000391946.2
|
KLC3
|
kinesin light chain 3 |
| chr19_+_45844018 | 2.35 |
ENST00000585434.1
|
KLC3
|
kinesin light chain 3 |
| chr19_-_14201507 | 2.03 |
ENST00000533683.2
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr9_+_34989638 | 2.02 |
ENST00000453597.3
ENST00000335998.3 ENST00000312316.5 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr11_-_66725837 | 1.93 |
ENST00000393958.2
ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC
|
pyruvate carboxylase |
| chr12_-_109125285 | 1.85 |
ENST00000552871.1
ENST00000261401.3 |
CORO1C
|
coronin, actin binding protein, 1C |
| chr7_+_100797726 | 1.79 |
ENST00000429457.1
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr3_-_124774802 | 1.78 |
ENST00000311127.4
|
HEG1
|
heart development protein with EGF-like domains 1 |
| chr1_-_93426998 | 1.78 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr8_-_144651024 | 1.73 |
ENST00000524906.1
ENST00000532862.1 ENST00000534459.1 |
MROH6
|
maestro heat-like repeat family member 6 |
| chr14_-_69445968 | 1.70 |
ENST00000438964.2
|
ACTN1
|
actinin, alpha 1 |
| chr12_+_70760056 | 1.70 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chrX_-_46618490 | 1.68 |
ENST00000328306.4
|
SLC9A7
|
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chr16_+_30194916 | 1.66 |
ENST00000570045.1
ENST00000565497.1 ENST00000570244.1 |
CORO1A
|
coronin, actin binding protein, 1A |
| chr19_-_14201776 | 1.66 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr2_+_30454390 | 1.65 |
ENST00000395323.3
ENST00000406087.1 ENST00000404397.1 |
LBH
|
limb bud and heart development |
| chr7_-_100860851 | 1.65 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr9_+_128509624 | 1.63 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr17_-_41623691 | 1.61 |
ENST00000545954.1
|
ETV4
|
ets variant 4 |
| chr19_-_51568324 | 1.60 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr15_+_63340775 | 1.60 |
ENST00000559281.1
ENST00000317516.7 |
TPM1
|
tropomyosin 1 (alpha) |
| chr15_-_91537723 | 1.59 |
ENST00000394249.3
ENST00000559811.1 ENST00000442656.2 ENST00000557905.1 ENST00000361919.3 |
PRC1
|
protein regulator of cytokinesis 1 |
| chr8_+_42752053 | 1.59 |
ENST00000307602.4
|
HOOK3
|
hook microtubule-tethering protein 3 |
| chr19_+_676385 | 1.59 |
ENST00000166139.4
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
| chr2_-_177502659 | 1.57 |
ENST00000295549.4
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr19_+_45844032 | 1.57 |
ENST00000589837.1
|
KLC3
|
kinesin light chain 3 |
| chr2_+_23608064 | 1.55 |
ENST00000486442.1
|
KLHL29
|
kelch-like family member 29 |
| chr11_-_66104237 | 1.55 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr4_+_8201091 | 1.53 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr2_-_224903995 | 1.52 |
ENST00000409304.1
ENST00000454956.1 ENST00000258405.4 |
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr15_-_75017711 | 1.51 |
ENST00000567032.1
ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr8_+_32405728 | 1.50 |
ENST00000523079.1
ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1
|
neuregulin 1 |
| chr9_+_128509663 | 1.49 |
ENST00000373489.5
ENST00000373483.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr11_-_62313090 | 1.48 |
ENST00000528508.1
ENST00000533365.1 |
AHNAK
|
AHNAK nucleoprotein |
| chr9_-_101017900 | 1.47 |
ENST00000375066.5
|
TBC1D2
|
TBC1 domain family, member 2 |
| chr15_+_63340734 | 1.46 |
ENST00000560959.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chrX_+_103173457 | 1.44 |
ENST00000419165.1
|
TMSB15B
|
thymosin beta 15B |
| chr18_+_33877654 | 1.42 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr17_-_36413133 | 1.42 |
ENST00000523089.1
ENST00000312412.4 ENST00000520237.1 |
RP11-1407O15.2
|
TBC1 domain family member 3 |
| chr19_+_54372639 | 1.37 |
ENST00000391769.2
|
MYADM
|
myeloid-associated differentiation marker |
| chr1_+_150521876 | 1.34 |
ENST00000369041.5
ENST00000271643.4 ENST00000538795.1 |
ADAMTSL4
AL356356.1
|
ADAMTS-like 4 Protein LOC100996516 |
| chr15_+_63340647 | 1.34 |
ENST00000404484.4
|
TPM1
|
tropomyosin 1 (alpha) |
| chr11_-_64546202 | 1.33 |
ENST00000377390.3
ENST00000227503.9 ENST00000377394.3 ENST00000422298.2 ENST00000334944.5 |
SF1
|
splicing factor 1 |
| chr1_-_183604794 | 1.33 |
ENST00000367534.1
ENST00000359856.6 ENST00000294742.6 |
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa |
| chr12_-_85306594 | 1.32 |
ENST00000266682.5
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr19_+_10400615 | 1.30 |
ENST00000221980.4
|
ICAM5
|
intercellular adhesion molecule 5, telencephalin |
| chr1_-_6321035 | 1.29 |
ENST00000377893.2
|
GPR153
|
G protein-coupled receptor 153 |
| chr18_-_658244 | 1.29 |
ENST00000585033.1
ENST00000323813.3 |
C18orf56
|
chromosome 18 open reading frame 56 |
| chr19_-_35992780 | 1.28 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr11_+_13690249 | 1.27 |
ENST00000532701.1
|
FAR1
|
fatty acyl CoA reductase 1 |
| chr20_+_44035200 | 1.27 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chrX_+_135229600 | 1.26 |
ENST00000370690.3
|
FHL1
|
four and a half LIM domains 1 |
| chrX_-_152939133 | 1.25 |
ENST00000370150.1
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr15_+_57884086 | 1.24 |
ENST00000380569.2
ENST00000380561.2 ENST00000574161.1 ENST00000572390.1 ENST00000396180.1 ENST00000380560.2 |
GCOM1
|
GRINL1A complex locus 1 |
| chr11_+_124609742 | 1.21 |
ENST00000284292.6
|
NRGN
|
neurogranin (protein kinase C substrate, RC3) |
| chrX_+_23925918 | 1.21 |
ENST00000379211.3
|
CXorf58
|
chromosome X open reading frame 58 |
| chr3_-_13921594 | 1.20 |
ENST00000285018.4
|
WNT7A
|
wingless-type MMTV integration site family, member 7A |
| chr19_-_45908292 | 1.19 |
ENST00000360957.5
ENST00000592134.1 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr6_-_31869769 | 1.19 |
ENST00000375527.2
|
ZBTB12
|
zinc finger and BTB domain containing 12 |
| chr9_-_34637806 | 1.19 |
ENST00000477726.1
|
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr10_-_134145321 | 1.19 |
ENST00000368625.4
ENST00000368619.3 ENST00000456004.1 ENST00000368620.2 |
STK32C
|
serine/threonine kinase 32C |
| chr6_-_109777128 | 1.18 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr7_+_100797678 | 1.18 |
ENST00000337619.5
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr19_-_3801789 | 1.17 |
ENST00000590849.1
ENST00000395045.2 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr10_+_124221036 | 1.17 |
ENST00000368984.3
|
HTRA1
|
HtrA serine peptidase 1 |
| chrX_-_152939252 | 1.17 |
ENST00000340888.3
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr1_-_150208291 | 1.16 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr12_+_66217911 | 1.16 |
ENST00000403681.2
|
HMGA2
|
high mobility group AT-hook 2 |
| chr11_-_66103932 | 1.14 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr21_-_16437126 | 1.13 |
ENST00000318948.4
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr9_+_34652164 | 1.12 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr6_-_2876744 | 1.12 |
ENST00000420981.2
|
RP11-420G6.4
|
RP11-420G6.4 |
| chr1_+_35246775 | 1.11 |
ENST00000373366.2
|
GJB3
|
gap junction protein, beta 3, 31kDa |
| chr7_-_32931623 | 1.11 |
ENST00000452926.1
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr2_+_204193149 | 1.11 |
ENST00000422511.2
|
ABI2
|
abl-interactor 2 |
| chr5_+_14143728 | 1.09 |
ENST00000344204.4
ENST00000537187.1 |
TRIO
|
trio Rho guanine nucleotide exchange factor |
| chr19_-_41859814 | 1.09 |
ENST00000221930.5
|
TGFB1
|
transforming growth factor, beta 1 |
| chr7_+_116593953 | 1.09 |
ENST00000397750.3
|
ST7-OT4
|
ST7 overlapping transcript 4 |
| chr16_-_58231782 | 1.08 |
ENST00000565188.1
ENST00000262506.3 |
CSNK2A2
|
casein kinase 2, alpha prime polypeptide |
| chr12_+_57482665 | 1.08 |
ENST00000300131.3
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr11_+_124609823 | 1.07 |
ENST00000412681.2
|
NRGN
|
neurogranin (protein kinase C substrate, RC3) |
| chr22_-_29137771 | 1.07 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr14_-_24610779 | 1.07 |
ENST00000560403.1
ENST00000419198.2 ENST00000216799.4 |
EMC9
|
ER membrane protein complex subunit 9 |
| chr5_+_150404904 | 1.07 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr9_+_137218362 | 1.07 |
ENST00000481739.1
|
RXRA
|
retinoid X receptor, alpha |
| chrX_-_48814278 | 1.06 |
ENST00000455452.1
|
OTUD5
|
OTU domain containing 5 |
| chr4_+_1873100 | 1.05 |
ENST00000508803.1
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr10_-_135150367 | 1.05 |
ENST00000368555.3
ENST00000252939.4 ENST00000368558.1 ENST00000368556.2 |
CALY
|
calcyon neuron-specific vesicular protein |
| chr8_-_127570603 | 1.05 |
ENST00000304916.3
|
FAM84B
|
family with sequence similarity 84, member B |
| chr16_+_67062996 | 1.03 |
ENST00000561924.2
|
CBFB
|
core-binding factor, beta subunit |
| chr17_-_41623716 | 1.03 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr10_+_11784360 | 1.02 |
ENST00000379215.4
ENST00000420401.1 |
ECHDC3
|
enoyl CoA hydratase domain containing 3 |
| chr18_+_55102917 | 1.02 |
ENST00000491143.2
|
ONECUT2
|
one cut homeobox 2 |
| chr2_+_191745560 | 1.02 |
ENST00000338435.4
|
GLS
|
glutaminase |
| chr16_+_1756162 | 1.01 |
ENST00000250894.4
ENST00000356010.5 |
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr2_+_204193129 | 1.01 |
ENST00000417864.1
|
ABI2
|
abl-interactor 2 |
| chr14_+_66974845 | 1.00 |
ENST00000459628.1
|
GPHN
|
gephyrin |
| chr15_+_42066740 | 1.00 |
ENST00000514566.1
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr10_+_99473455 | 0.99 |
ENST00000285605.6
|
MARVELD1
|
MARVEL domain containing 1 |
| chr6_+_35995552 | 0.99 |
ENST00000468133.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr12_-_49351228 | 0.99 |
ENST00000541959.1
ENST00000447318.2 |
ARF3
|
ADP-ribosylation factor 3 |
| chr19_-_15311713 | 0.99 |
ENST00000601011.1
ENST00000263388.2 |
NOTCH3
|
notch 3 |
| chrX_-_154299501 | 0.98 |
ENST00000369476.3
ENST00000369484.3 |
MTCP1
CMC4
|
mature T-cell proliferation 1 C-x(9)-C motif containing 4 |
| chr18_-_47340297 | 0.98 |
ENST00000586485.1
ENST00000587994.1 ENST00000586100.1 ENST00000285093.10 |
ACAA2
|
acetyl-CoA acyltransferase 2 |
| chr16_+_56598961 | 0.97 |
ENST00000219162.3
|
MT4
|
metallothionein 4 |
| chr8_-_30515693 | 0.97 |
ENST00000355904.4
|
GTF2E2
|
general transcription factor IIE, polypeptide 2, beta 34kDa |
| chr21_-_16437255 | 0.96 |
ENST00000400199.1
ENST00000400202.1 |
NRIP1
|
nuclear receptor interacting protein 1 |
| chr7_+_143013198 | 0.96 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr6_-_144329531 | 0.96 |
ENST00000429150.1
ENST00000392309.1 ENST00000416623.1 ENST00000392307.1 |
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chrX_-_48814810 | 0.96 |
ENST00000376488.3
ENST00000396743.3 ENST00000156084.4 |
OTUD5
|
OTU domain containing 5 |
| chr21_-_47648665 | 0.96 |
ENST00000450351.1
ENST00000522411.1 ENST00000356396.4 ENST00000397728.3 ENST00000457828.2 |
LSS
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr15_+_42066632 | 0.95 |
ENST00000457542.2
ENST00000221214.6 ENST00000260357.7 ENST00000456763.2 |
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr18_+_12308231 | 0.95 |
ENST00000590103.1
ENST00000591909.1 ENST00000586653.1 ENST00000592683.1 ENST00000590967.1 ENST00000591208.1 ENST00000591463.1 |
TUBB6
|
tubulin, beta 6 class V |
| chr2_+_54683419 | 0.95 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr19_-_2050852 | 0.95 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr17_-_7297519 | 0.94 |
ENST00000576362.1
ENST00000571078.1 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr1_+_6052700 | 0.94 |
ENST00000378092.1
ENST00000445501.1 |
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr20_-_22565101 | 0.93 |
ENST00000419308.2
|
FOXA2
|
forkhead box A2 |
| chr6_-_112194484 | 0.92 |
ENST00000518295.1
ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr8_-_124408652 | 0.92 |
ENST00000287394.5
|
ATAD2
|
ATPase family, AAA domain containing 2 |
| chr19_-_40791211 | 0.92 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr17_-_7297833 | 0.92 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr11_+_67070919 | 0.91 |
ENST00000308127.4
ENST00000308298.7 |
SSH3
|
slingshot protein phosphatase 3 |
| chr14_-_69446034 | 0.91 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr7_-_143105941 | 0.90 |
ENST00000275815.3
|
EPHA1
|
EPH receptor A1 |
| chr1_-_38471156 | 0.90 |
ENST00000373016.3
|
FHL3
|
four and a half LIM domains 3 |
| chr14_-_69445793 | 0.90 |
ENST00000538545.2
ENST00000394419.4 |
ACTN1
|
actinin, alpha 1 |
| chr19_-_663277 | 0.89 |
ENST00000292363.5
|
RNF126
|
ring finger protein 126 |
| chr14_-_55493763 | 0.89 |
ENST00000455555.1
ENST00000360586.3 ENST00000421192.1 ENST00000420358.2 |
WDHD1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr1_-_204121102 | 0.89 |
ENST00000367202.4
|
ETNK2
|
ethanolamine kinase 2 |
| chr15_+_40674920 | 0.88 |
ENST00000416151.2
ENST00000249776.8 |
KNSTRN
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr16_+_28986085 | 0.88 |
ENST00000565975.1
ENST00000311008.11 ENST00000323081.8 ENST00000334536.8 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr5_-_115177247 | 0.88 |
ENST00000500945.2
|
ATG12
|
autophagy related 12 |
| chr2_+_228337079 | 0.87 |
ENST00000409315.1
ENST00000373671.3 ENST00000409171.1 |
AGFG1
|
ArfGAP with FG repeats 1 |
| chr14_+_100070869 | 0.87 |
ENST00000502101.2
|
RP11-543C4.1
|
RP11-543C4.1 |
| chr14_+_66975213 | 0.86 |
ENST00000543237.1
ENST00000305960.9 |
GPHN
|
gephyrin |
| chr17_-_41623075 | 0.86 |
ENST00000545089.1
|
ETV4
|
ets variant 4 |
| chr20_+_306177 | 0.86 |
ENST00000544632.1
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chrX_+_117480036 | 0.86 |
ENST00000371822.5
ENST00000254029.3 ENST00000371825.3 |
WDR44
|
WD repeat domain 44 |
| chr19_+_30302805 | 0.85 |
ENST00000262643.3
ENST00000575243.1 ENST00000357943.5 |
CCNE1
|
cyclin E1 |
| chr15_+_40674963 | 0.85 |
ENST00000448395.2
|
KNSTRN
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr19_+_16178317 | 0.84 |
ENST00000344824.6
ENST00000538887.1 |
TPM4
|
tropomyosin 4 |
| chr5_-_140998616 | 0.84 |
ENST00000389054.3
ENST00000398562.2 ENST00000389057.5 ENST00000398566.3 ENST00000398557.4 ENST00000253811.6 |
DIAPH1
|
diaphanous-related formin 1 |
| chr11_+_67071050 | 0.84 |
ENST00000376757.5
|
SSH3
|
slingshot protein phosphatase 3 |
| chr16_+_3070313 | 0.84 |
ENST00000326577.4
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr16_+_28986134 | 0.84 |
ENST00000352260.7
|
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr12_+_1099675 | 0.84 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr3_+_50192537 | 0.83 |
ENST00000002829.3
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr16_+_11038403 | 0.83 |
ENST00000409552.3
|
CLEC16A
|
C-type lectin domain family 16, member A |
| chr3_+_50192499 | 0.83 |
ENST00000413852.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chrX_-_154033793 | 0.83 |
ENST00000369534.3
ENST00000413259.3 |
MPP1
|
membrane protein, palmitoylated 1, 55kDa |
| chr1_+_3388181 | 0.82 |
ENST00000418137.1
ENST00000413250.2 |
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr12_-_53625958 | 0.82 |
ENST00000327550.3
ENST00000546717.1 ENST00000425354.2 ENST00000394426.1 |
RARG
|
retinoic acid receptor, gamma |
| chr19_-_38397285 | 0.82 |
ENST00000303868.5
|
WDR87
|
WD repeat domain 87 |
| chr2_+_220306745 | 0.82 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr19_-_14629224 | 0.82 |
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr1_+_26606608 | 0.81 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr5_-_176923846 | 0.81 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr15_+_80696666 | 0.80 |
ENST00000303329.4
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr19_-_13617247 | 0.80 |
ENST00000573710.2
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr19_-_41196534 | 0.80 |
ENST00000252891.4
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr4_-_82393009 | 0.79 |
ENST00000436139.2
|
RASGEF1B
|
RasGEF domain family, member 1B |
| chr8_+_59465728 | 0.79 |
ENST00000260130.4
ENST00000422546.2 ENST00000447182.2 ENST00000413219.2 ENST00000424270.2 ENST00000523483.1 ENST00000520168.1 |
SDCBP
|
syndecan binding protein (syntenin) |
| chr1_+_24645865 | 0.79 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr19_-_44174330 | 0.79 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr6_+_99282570 | 0.79 |
ENST00000328345.5
|
POU3F2
|
POU class 3 homeobox 2 |
| chr8_-_63951730 | 0.79 |
ENST00000260118.6
|
GGH
|
gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) |
| chrX_-_51239425 | 0.79 |
ENST00000375992.3
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr6_+_35995488 | 0.78 |
ENST00000229795.3
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr1_-_85155939 | 0.78 |
ENST00000603677.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr1_+_24645807 | 0.77 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr14_+_55033815 | 0.77 |
ENST00000554335.1
|
SAMD4A
|
sterile alpha motif domain containing 4A |
Network of associatons between targets according to the STRING database.
First level regulatory network of EGR1_EGR4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 1.4 | 4.2 | GO:0002605 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) |
| 1.1 | 3.4 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 1.1 | 3.2 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 1.0 | 2.9 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.7 | 6.4 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.6 | 1.9 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.6 | 1.9 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.6 | 1.8 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.6 | 1.7 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.6 | 4.4 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.5 | 7.1 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.5 | 6.7 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.5 | 1.5 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.5 | 11.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.5 | 2.5 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.5 | 1.5 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.5 | 1.5 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.4 | 3.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.4 | 3.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.4 | 11.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.4 | 1.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.4 | 3.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.4 | 1.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.4 | 6.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 1.2 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.4 | 1.5 | GO:0009644 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.4 | 1.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.4 | 1.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.4 | 1.5 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.4 | 1.5 | GO:2000685 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.4 | 1.5 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.4 | 1.8 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.4 | 1.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.4 | 1.1 | GO:0075528 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.3 | 1.7 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.3 | 1.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 | 1.7 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.3 | 1.7 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.3 | 3.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 1.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 1.3 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.3 | 1.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.3 | 0.9 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.3 | 1.5 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.3 | 1.7 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.3 | 0.3 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.3 | 2.0 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.3 | 1.9 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.3 | 1.7 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.3 | 0.8 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.3 | 1.5 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.3 | 3.8 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 0.5 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.2 | 0.7 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.2 | 1.2 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.2 | 1.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 0.9 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 1.8 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 0.4 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.2 | 1.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.2 | 0.6 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 0.6 | GO:0048320 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) axial mesoderm formation(GO:0048320) |
| 0.2 | 0.6 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.2 | 1.7 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.2 | 0.8 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.2 | 0.6 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.2 | 0.6 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.2 | 0.6 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.2 | 0.2 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.2 | 0.6 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.2 | 1.0 | GO:0072103 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.2 | 0.4 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.2 | 2.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 | 2.1 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.2 | 0.8 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.2 | 0.9 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 0.6 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.2 | 0.5 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.2 | 0.5 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.2 | 1.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.2 | 0.5 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 1.6 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.2 | 0.5 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.2 | 0.7 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 0.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 | 0.7 | GO:0072300 | positive regulation of metanephric glomerulus development(GO:0072300) |
| 0.2 | 0.8 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.2 | 1.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 1.0 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.2 | 0.5 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.2 | 1.0 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.2 | 0.5 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.2 | 2.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 0.8 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.2 | 0.6 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.2 | 2.4 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 0.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 0.5 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.2 | 4.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 0.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 0.8 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 0.9 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.4 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 | 2.5 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.1 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 1.5 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 0.6 | GO:1901895 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.1 | 0.6 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.1 | 0.7 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 0.4 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.3 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.4 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.1 | 0.5 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 1.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.8 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.7 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 0.7 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.7 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.8 | GO:1901906 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 1.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 1.3 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.1 | 0.4 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.9 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.1 | 2.4 | GO:0060973 | cell migration involved in heart development(GO:0060973) |
| 0.1 | 0.7 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.1 | 0.2 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 1.9 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.7 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.3 | GO:0060585 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.6 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.8 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 0.7 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 1.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.3 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 1.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 1.0 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 0.6 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 1.0 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.4 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.3 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.1 | 0.5 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 2.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.3 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.1 | 0.6 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.9 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.3 | GO:0061308 | cardiac neural crest cell differentiation involved in heart development(GO:0061307) cardiac neural crest cell development involved in heart development(GO:0061308) |
| 0.1 | 0.2 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.1 | 0.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.4 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) lung growth(GO:0060437) |
| 0.1 | 1.7 | GO:0090520 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 1.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.3 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.7 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 1.0 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 0.2 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.1 | 1.5 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 1.6 | GO:0000022 | mitotic spindle elongation(GO:0000022) |
| 0.1 | 0.3 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.1 | 0.7 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.2 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.1 | 1.0 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 | 0.4 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.2 | GO:0048382 | mesodermal cell migration(GO:0008078) forebrain dorsal/ventral pattern formation(GO:0021798) mesendoderm development(GO:0048382) |
| 0.1 | 0.6 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.1 | 0.7 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 1.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 1.0 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.4 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.3 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.6 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.3 | GO:0019087 | transformation of host cell by virus(GO:0019087) renal water absorption(GO:0070295) |
| 0.1 | 0.9 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.8 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.4 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.3 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.5 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.1 | 4.4 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.7 | GO:0007135 | meiosis II(GO:0007135) |
| 0.1 | 0.3 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 1.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.1 | GO:0030820 | regulation of cyclic nucleotide catabolic process(GO:0030805) regulation of cAMP catabolic process(GO:0030820) regulation of purine nucleotide catabolic process(GO:0033121) |
| 0.1 | 0.5 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 1.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.3 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.1 | GO:0090178 | regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) |
| 0.1 | 0.4 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.6 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 3.1 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 0.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.8 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 1.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 1.0 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.9 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.7 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.1 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 8.0 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 0.4 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 0.3 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.5 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.7 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 0.5 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.3 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.1 | 0.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.3 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.6 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.1 | 0.3 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.1 | 1.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.6 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 0.7 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.7 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 1.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.5 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 1.5 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.5 | GO:1905245 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.0 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 | 0.4 | GO:0060318 | regulation of definitive erythrocyte differentiation(GO:0010724) definitive erythrocyte differentiation(GO:0060318) |
| 0.0 | 0.1 | GO:0019520 | pentose-phosphate shunt, oxidative branch(GO:0009051) aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.2 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 1.5 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.2 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.0 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.0 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 1.2 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.0 | 2.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.6 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.6 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.7 | GO:0060444 | branching involved in mammary gland duct morphogenesis(GO:0060444) |
| 0.0 | 0.7 | GO:0036119 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.4 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.8 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.4 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.4 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.3 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.7 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0060592 | mammary gland formation(GO:0060592) |
| 0.0 | 0.2 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.3 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.0 | 0.4 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 | 0.3 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.7 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.0 | 0.3 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.1 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.6 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.7 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.6 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.7 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.4 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.5 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 0.9 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.9 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.4 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.5 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.4 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 1.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 2.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.8 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.4 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.3 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.7 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.7 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.0 | 0.3 | GO:1903317 | regulation of protein processing(GO:0070613) regulation of protein maturation(GO:1903317) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 0.6 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.5 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 0.1 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.0 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 1.1 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.2 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) positive regulation of transcription by glucose(GO:0046016) |
| 0.0 | 0.1 | GO:0070142 | synaptic vesicle budding(GO:0070142) |
| 0.0 | 0.3 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.2 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.4 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.8 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.2 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.8 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0044557 | relaxation of smooth muscle(GO:0044557) |
| 0.0 | 0.4 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.5 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 0.7 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.3 | GO:0031958 | corticosteroid receptor signaling pathway(GO:0031958) glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.6 | GO:0030818 | negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.3 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) |
| 0.0 | 0.4 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.2 | GO:0006388 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.8 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.0 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.3 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.8 | GO:0072431 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 | 1.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.6 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.6 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.4 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.6 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.0 | 0.1 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.0 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.6 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.6 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.9 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.6 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.1 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.3 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.2 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.4 | GO:0071174 | mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.2 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 0.2 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.0 | GO:0044026 | DNA hypermethylation(GO:0044026) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 0.8 | GO:0000806 | Y chromosome(GO:0000806) |
| 0.6 | 1.2 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.6 | 9.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.4 | 3.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.4 | 6.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 0.7 | GO:0036019 | endolysosome(GO:0036019) endolysosome membrane(GO:0036020) |
| 0.3 | 2.7 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.3 | 4.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 0.9 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.3 | 0.9 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.3 | 5.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 1.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 0.7 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.2 | 1.6 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 3.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 3.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 0.6 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.2 | 12.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 2.5 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 1.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 0.5 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.2 | 1.8 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 1.0 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 0.7 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.9 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.9 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 1.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.5 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 3.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 1.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.5 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 1.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.8 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.6 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.4 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.1 | 1.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 3.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 3.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.5 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.2 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 8.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 1.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.4 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 1.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 7.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.6 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 1.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.6 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.3 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.5 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 1.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.7 | GO:0032991 | macromolecular complex(GO:0032991) |
| 0.0 | 1.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 4.6 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 1.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 3.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.1 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.4 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 3.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.7 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 3.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 1.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 5.2 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 2.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.0 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.5 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 7.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.8 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.8 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 1.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.5 | GO:0031253 | cell projection membrane(GO:0031253) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.0 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0005903 | brush border(GO:0005903) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.9 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 1.4 | 4.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.6 | 1.9 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.6 | 2.3 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.6 | 0.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.5 | 3.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.5 | 1.5 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.4 | 1.3 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.4 | 1.5 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.3 | 1.7 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.3 | 1.7 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.3 | 7.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 1.0 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.3 | 1.0 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.3 | 1.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 3.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 1.5 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.3 | 1.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.3 | 9.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 0.8 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 1.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 0.8 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.3 | 0.8 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.3 | 0.8 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 4.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 2.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 1.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 4.8 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 1.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 0.6 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.2 | 0.6 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.2 | 0.6 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.2 | 1.9 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.2 | 8.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 0.9 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.2 | 1.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 0.6 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.2 | 1.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.2 | 0.7 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 1.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 0.5 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.2 | 0.5 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.2 | 0.9 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.5 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.2 | 0.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 0.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 2.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.2 | 0.9 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 0.6 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.2 | 0.8 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 3.5 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.4 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.9 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.4 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 3.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.6 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.4 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.7 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 1.9 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 2.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.8 | GO:0052843 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.8 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.1 | 0.5 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.6 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 1.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 1.1 | GO:0004673 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) |
| 0.1 | 0.6 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.7 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 4.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.3 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 1.0 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.1 | 1.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 2.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 1.0 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.1 | 1.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.3 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.1 | 1.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 1.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.7 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.3 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 4.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 4.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.9 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 2.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 1.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.3 | GO:0070364 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.1 | 0.5 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.4 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 0.5 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.3 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 1.2 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 0.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 5.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.3 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 1.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 2.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.3 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.3 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 0.4 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 1.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.8 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.4 | GO:0042835 | BRE binding(GO:0042835) |
| 0.1 | 1.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.3 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 2.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 8.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 3.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.0 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.1 | 0.6 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 1.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 2.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 3.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.7 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 2.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.3 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.6 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 5.2 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 4.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 1.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 2.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.9 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 10.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 3.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 1.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.8 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 3.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.3 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 1.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.8 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 3.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.9 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.8 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.4 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.3 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 0.4 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.7 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 2.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 1.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.0 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 2.2 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.0 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 1.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.9 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 13.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 8.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 3.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 8.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 2.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 4.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 0.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 5.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 6.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 6.2 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 8.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 1.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 2.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 5.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.8 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 0.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 4.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 3.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 0.9 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 3.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.3 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 4.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 6.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 3.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 5.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 3.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 5.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 0.3 | REACTOME CELL CELL COMMUNICATION | Genes involved in Cell-Cell communication |
| 0.1 | 2.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 2.9 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 2.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 4.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 5.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.3 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 1.1 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.8 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 0.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 0.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 1.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 2.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 3.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 1.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 2.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 2.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 0.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 3.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.9 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 1.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 2.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.7 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 2.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.8 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 1.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.6 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.1 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.0 | 0.8 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |