Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
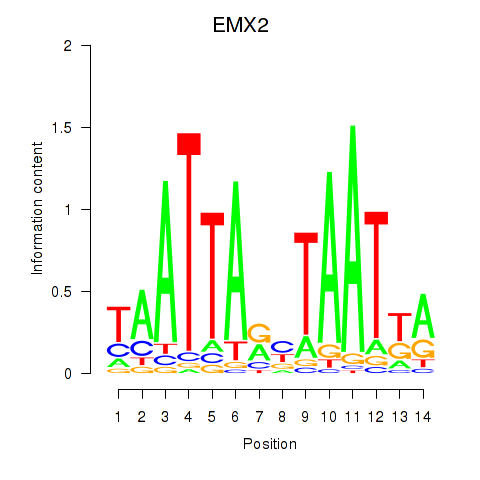
Results for EMX2
Z-value: 0.52
Transcription factors associated with EMX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX2
|
ENSG00000170370.10 | empty spiracles homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX2 | hg19_v2_chr10_+_119301928_119301955 | -0.05 | 8.1e-01 | Click! |
Activity profile of EMX2 motif
Sorted Z-values of EMX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_63638339 | 3.70 |
ENST00000343837.3
ENST00000469440.1 |
SNTN
|
sentan, cilia apical structure protein |
| chr4_-_100356551 | 2.83 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr6_-_33041378 | 2.21 |
ENST00000428995.1
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr4_-_100356291 | 1.95 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr2_+_228736335 | 1.90 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr11_-_26593779 | 1.71 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr21_-_43735628 | 1.53 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr5_-_137475071 | 1.38 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr11_-_63376013 | 1.38 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr11_-_26593677 | 1.38 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr11_-_26593649 | 1.32 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr19_-_41388657 | 1.32 |
ENST00000301146.4
ENST00000291764.3 |
CYP2A7
|
cytochrome P450, family 2, subfamily A, polypeptide 7 |
| chr21_-_43735446 | 1.32 |
ENST00000398431.2
|
TFF3
|
trefoil factor 3 (intestinal) |
| chr12_-_68696652 | 1.22 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr16_-_28634874 | 1.20 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr12_-_71533055 | 1.15 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chr3_+_113616317 | 1.13 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr8_-_133687813 | 1.03 |
ENST00000250173.1
ENST00000519595.1 |
LRRC6
|
leucine rich repeat containing 6 |
| chr1_-_86848760 | 1.03 |
ENST00000460698.2
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr8_-_133687778 | 0.98 |
ENST00000518642.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr1_+_36549676 | 0.90 |
ENST00000207457.3
|
TEKT2
|
tektin 2 (testicular) |
| chr1_+_79115503 | 0.87 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr6_-_52705641 | 0.86 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr1_+_170904612 | 0.86 |
ENST00000367759.4
ENST00000367758.3 |
MROH9
|
maestro heat-like repeat family member 9 |
| chr3_+_130745688 | 0.85 |
ENST00000510769.1
ENST00000429253.2 ENST00000356918.4 ENST00000510688.1 ENST00000511262.1 ENST00000383366.4 |
NEK11
|
NIMA-related kinase 11 |
| chr6_+_27833034 | 0.83 |
ENST00000357320.2
|
HIST1H2AL
|
histone cluster 1, H2al |
| chrX_+_36254051 | 0.76 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr22_-_51222070 | 0.73 |
ENST00000395593.3
ENST00000395598.3 ENST00000435118.1 ENST00000395591.1 ENST00000395595.3 |
RABL2B
|
RAB, member of RAS oncogene family-like 2B |
| chr5_-_111093167 | 0.73 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr11_+_27076764 | 0.69 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr3_-_122512619 | 0.66 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr6_-_109761707 | 0.64 |
ENST00000520723.1
ENST00000518648.1 ENST00000417394.2 |
PPIL6
|
peptidylprolyl isomerase (cyclophilin)-like 6 |
| chr12_-_10282836 | 0.63 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr2_-_99279928 | 0.63 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr11_+_66276550 | 0.60 |
ENST00000419755.3
|
CTD-3074O7.11
|
Bardet-Biedl syndrome 1 protein |
| chr14_+_74034310 | 0.60 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr5_-_111092930 | 0.56 |
ENST00000257435.7
|
NREP
|
neuronal regeneration related protein |
| chr7_+_90338712 | 0.55 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr5_-_111312622 | 0.53 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr19_+_58144529 | 0.52 |
ENST00000347302.3
ENST00000254182.7 ENST00000391703.3 ENST00000541801.1 ENST00000299871.5 ENST00000544273.1 |
ZNF211
|
zinc finger protein 211 |
| chr6_-_76072719 | 0.52 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr13_-_70682590 | 0.51 |
ENST00000377844.4
|
KLHL1
|
kelch-like family member 1 |
| chr19_+_49199209 | 0.51 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr6_+_150690028 | 0.51 |
ENST00000229447.5
ENST00000344419.3 |
IYD
|
iodotyrosine deiodinase |
| chr1_-_35450897 | 0.50 |
ENST00000373337.3
|
ZMYM6NB
|
ZMYM6 neighbor |
| chrX_+_144908928 | 0.50 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr12_+_9980113 | 0.50 |
ENST00000537723.1
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr15_+_84115868 | 0.49 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr5_-_111093081 | 0.48 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr4_-_25865159 | 0.48 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr4_+_74702214 | 0.48 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr4_+_56212270 | 0.48 |
ENST00000264228.4
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr4_-_138453559 | 0.47 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr8_+_110552831 | 0.46 |
ENST00000530629.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr6_-_134639180 | 0.46 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr14_+_51338816 | 0.46 |
ENST00000353130.1
ENST00000337334.2 ENST00000395752.1 |
ABHD12B
|
abhydrolase domain containing 12B |
| chr12_-_10282742 | 0.46 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr12_+_21207503 | 0.45 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr16_-_28937027 | 0.45 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr12_-_10282681 | 0.45 |
ENST00000533022.1
|
CLEC7A
|
C-type lectin domain family 7, member A |
| chr19_+_50016610 | 0.43 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr16_+_69599861 | 0.42 |
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr15_-_57025759 | 0.42 |
ENST00000267807.7
|
ZNF280D
|
zinc finger protein 280D |
| chr19_-_35264089 | 0.41 |
ENST00000588760.1
ENST00000329285.8 ENST00000587354.2 |
ZNF599
|
zinc finger protein 599 |
| chrX_+_55744228 | 0.40 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr5_-_41213607 | 0.39 |
ENST00000337836.5
ENST00000433294.1 |
C6
|
complement component 6 |
| chr14_+_102276132 | 0.38 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr3_-_130745571 | 0.37 |
ENST00000514044.1
ENST00000264992.3 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr6_-_26235206 | 0.37 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr12_-_122985067 | 0.36 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr4_-_186877806 | 0.36 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chrX_+_55744166 | 0.36 |
ENST00000374941.4
ENST00000414239.1 |
RRAGB
|
Ras-related GTP binding B |
| chr6_+_116421976 | 0.35 |
ENST00000319550.4
ENST00000419791.1 |
NT5DC1
|
5'-nucleotidase domain containing 1 |
| chr4_+_56212505 | 0.35 |
ENST00000505210.1
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr15_-_75748115 | 0.35 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr7_-_73153178 | 0.35 |
ENST00000437775.2
ENST00000222800.3 |
ABHD11
|
abhydrolase domain containing 11 |
| chr4_-_186877502 | 0.35 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr12_-_100656134 | 0.34 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr11_-_128894053 | 0.33 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr3_+_132316081 | 0.33 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr16_-_28374829 | 0.33 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr8_+_110552337 | 0.32 |
ENST00000337573.5
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr3_-_49466686 | 0.31 |
ENST00000273598.3
ENST00000436744.2 |
NICN1
|
nicolin 1 |
| chrX_+_77166172 | 0.31 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr3_+_140981456 | 0.31 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr21_-_43346790 | 0.30 |
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chr4_-_168155169 | 0.30 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr5_+_112312416 | 0.30 |
ENST00000389063.2
|
DCP2
|
decapping mRNA 2 |
| chr8_-_144660771 | 0.29 |
ENST00000449291.2
|
NAPRT1
|
nicotinate phosphoribosyltransferase domain containing 1 |
| chr16_+_31885079 | 0.29 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr5_+_140514782 | 0.29 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr8_+_67687413 | 0.28 |
ENST00000521960.1
ENST00000522398.1 ENST00000522629.1 ENST00000520976.1 ENST00000396596.1 |
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr1_-_67142710 | 0.28 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr2_-_62733476 | 0.28 |
ENST00000335390.5
|
TMEM17
|
transmembrane protein 17 |
| chr5_-_55412774 | 0.28 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr1_-_54405773 | 0.27 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr16_+_15489603 | 0.27 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr12_-_58220078 | 0.27 |
ENST00000549039.1
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr12_+_94071341 | 0.27 |
ENST00000542893.2
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr6_+_28092338 | 0.27 |
ENST00000340487.4
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr1_+_174844645 | 0.26 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr11_+_10476851 | 0.26 |
ENST00000396553.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr2_-_207024233 | 0.25 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr15_-_78913628 | 0.24 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr19_-_14945933 | 0.24 |
ENST00000322301.3
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5 |
| chr12_+_94071129 | 0.24 |
ENST00000552983.1
ENST00000332896.3 ENST00000552033.1 ENST00000548483.1 |
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr1_+_159931002 | 0.24 |
ENST00000443364.1
ENST00000423943.1 |
RP11-48O20.4
|
long intergenic non-protein coding RNA 1133 |
| chr2_+_56179262 | 0.24 |
ENST00000606639.1
|
RP11-481J13.1
|
RP11-481J13.1 |
| chr3_-_186288097 | 0.23 |
ENST00000446782.1
|
TBCCD1
|
TBCC domain containing 1 |
| chr9_-_5833027 | 0.23 |
ENST00000339450.5
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr8_-_38008783 | 0.23 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr16_+_28648975 | 0.23 |
ENST00000529716.1
|
NPIPB8
|
nuclear pore complex interacting protein family, member B8 |
| chr16_+_28763108 | 0.23 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr20_-_29978383 | 0.22 |
ENST00000339144.3
ENST00000376321.3 |
DEFB119
|
defensin, beta 119 |
| chr7_-_44887620 | 0.22 |
ENST00000349299.3
ENST00000521529.1 ENST00000308153.4 ENST00000350771.3 ENST00000222690.6 ENST00000381124.5 ENST00000437072.1 ENST00000446531.1 |
H2AFV
|
H2A histone family, member V |
| chr1_-_170043709 | 0.22 |
ENST00000367767.1
ENST00000361580.2 ENST00000538366.1 |
KIFAP3
|
kinesin-associated protein 3 |
| chr4_-_100140331 | 0.22 |
ENST00000407820.2
ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr12_-_91574142 | 0.21 |
ENST00000547937.1
|
DCN
|
decorin |
| chr1_-_190446759 | 0.21 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr8_-_143859197 | 0.21 |
ENST00000395192.2
|
LYNX1
|
Ly6/neurotoxin 1 |
| chr12_+_9980069 | 0.21 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr11_-_124981475 | 0.21 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr17_-_4938712 | 0.21 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr4_+_41540160 | 0.20 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr11_+_122753391 | 0.19 |
ENST00000307257.6
ENST00000227349.2 |
C11orf63
|
chromosome 11 open reading frame 63 |
| chr1_+_233765353 | 0.19 |
ENST00000366620.1
|
KCNK1
|
potassium channel, subfamily K, member 1 |
| chr9_-_99540328 | 0.19 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr8_+_98900132 | 0.19 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chrX_+_105412290 | 0.18 |
ENST00000357175.2
ENST00000337685.2 |
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1 |
| chr5_-_58571935 | 0.18 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr6_+_30585486 | 0.18 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr9_+_71986182 | 0.18 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr1_-_13390765 | 0.18 |
ENST00000357367.2
|
PRAMEF8
|
PRAME family member 8 |
| chr5_+_81575281 | 0.18 |
ENST00000380167.4
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr17_-_42100474 | 0.17 |
ENST00000585950.1
ENST00000592127.1 ENST00000589334.1 |
TMEM101
|
transmembrane protein 101 |
| chr7_+_138145076 | 0.17 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr8_+_24151553 | 0.17 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chrX_+_130192318 | 0.17 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr12_+_10365404 | 0.16 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr9_+_71944241 | 0.16 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr6_+_6588316 | 0.16 |
ENST00000379953.2
|
LY86
|
lymphocyte antigen 86 |
| chr3_+_148508845 | 0.16 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr1_-_85870177 | 0.16 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr8_-_101571933 | 0.16 |
ENST00000520311.1
|
ANKRD46
|
ankyrin repeat domain 46 |
| chr11_-_327537 | 0.16 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr2_-_179343226 | 0.15 |
ENST00000434643.2
|
FKBP7
|
FK506 binding protein 7 |
| chr16_-_4817129 | 0.15 |
ENST00000545009.1
ENST00000219478.6 |
ZNF500
|
zinc finger protein 500 |
| chr3_+_40518599 | 0.15 |
ENST00000314686.5
ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619
|
zinc finger protein 619 |
| chr1_+_117544366 | 0.15 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr3_-_58523010 | 0.15 |
ENST00000459701.2
ENST00000302819.5 |
ACOX2
|
acyl-CoA oxidase 2, branched chain |
| chr15_-_41166414 | 0.15 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr2_+_44001172 | 0.15 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr7_-_140482926 | 0.15 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr5_+_178977546 | 0.15 |
ENST00000319449.4
ENST00000377001.2 |
RUFY1
|
RUN and FYVE domain containing 1 |
| chr13_+_108921977 | 0.14 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr17_-_39324424 | 0.14 |
ENST00000391356.2
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr15_+_77712993 | 0.14 |
ENST00000336216.4
ENST00000381714.3 ENST00000558651.1 |
HMG20A
|
high mobility group 20A |
| chr9_+_75766763 | 0.14 |
ENST00000456643.1
ENST00000415424.1 |
ANXA1
|
annexin A1 |
| chr1_-_19615744 | 0.14 |
ENST00000361640.4
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr4_-_120243545 | 0.14 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr2_-_197226875 | 0.13 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr15_+_64680003 | 0.13 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr15_-_65903407 | 0.13 |
ENST00000395644.4
ENST00000567744.1 ENST00000568573.1 ENST00000562830.1 ENST00000569491.1 ENST00000561769.1 |
VWA9
|
von Willebrand factor A domain containing 9 |
| chr6_+_110501621 | 0.13 |
ENST00000368930.1
ENST00000307731.1 |
CDC40
|
cell division cycle 40 |
| chr2_+_120687335 | 0.13 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chrX_+_18443703 | 0.13 |
ENST00000379996.3
|
CDKL5
|
cyclin-dependent kinase-like 5 |
| chr15_-_78913521 | 0.13 |
ENST00000326828.5
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr3_+_39424828 | 0.12 |
ENST00000273158.4
ENST00000431510.1 |
SLC25A38
|
solute carrier family 25, member 38 |
| chr4_-_68411275 | 0.12 |
ENST00000273853.6
|
CENPC
|
centromere protein C |
| chr8_-_57123815 | 0.12 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr16_+_22524844 | 0.12 |
ENST00000538606.1
ENST00000424340.1 ENST00000517539.1 ENST00000528249.1 |
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr14_-_90420862 | 0.12 |
ENST00000556005.1
ENST00000555872.1 |
EFCAB11
|
EF-hand calcium binding domain 11 |
| chr19_+_15852203 | 0.12 |
ENST00000305892.1
|
OR10H3
|
olfactory receptor, family 10, subfamily H, member 3 |
| chr3_+_32993065 | 0.12 |
ENST00000330953.5
|
CCR4
|
chemokine (C-C motif) receptor 4 |
| chr20_+_15177480 | 0.12 |
ENST00000402914.1
|
MACROD2
|
MACRO domain containing 2 |
| chr10_-_27529486 | 0.11 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr5_+_55147205 | 0.11 |
ENST00000396836.2
ENST00000396834.1 ENST00000447346.2 ENST00000359040.5 |
IL31RA
|
interleukin 31 receptor A |
| chrX_+_36053908 | 0.11 |
ENST00000378660.2
|
CHDC2
|
calponin homology domain containing 2 |
| chr4_+_113568207 | 0.11 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr9_-_116065551 | 0.11 |
ENST00000297894.5
|
RNF183
|
ring finger protein 183 |
| chr2_+_234826016 | 0.11 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr6_+_110501344 | 0.11 |
ENST00000368932.1
|
CDC40
|
cell division cycle 40 |
| chr13_+_103459704 | 0.11 |
ENST00000602836.1
|
BIVM-ERCC5
|
BIVM-ERCC5 readthrough |
| chr9_-_28670283 | 0.11 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr12_+_59989918 | 0.11 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr17_+_7239821 | 0.11 |
ENST00000158762.3
ENST00000570457.2 |
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr22_+_45725524 | 0.11 |
ENST00000405548.3
|
FAM118A
|
family with sequence similarity 118, member A |
| chr6_-_9933500 | 0.11 |
ENST00000492169.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr16_+_11038345 | 0.10 |
ENST00000409790.1
|
CLEC16A
|
C-type lectin domain family 16, member A |
| chr14_+_55493920 | 0.10 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr10_+_51187938 | 0.10 |
ENST00000311663.5
|
FAM21D
|
family with sequence similarity 21, member D |
| chr1_-_92952433 | 0.10 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr2_+_32502952 | 0.10 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr15_+_75970672 | 0.10 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr3_+_195413160 | 0.10 |
ENST00000599448.1
|
LINC00969
|
long intergenic non-protein coding RNA 969 |
| chr3_+_38080691 | 0.09 |
ENST00000308059.6
ENST00000346219.3 ENST00000452631.2 |
DLEC1
|
deleted in lung and esophageal cancer 1 |
| chr11_-_102714534 | 0.09 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chrX_-_9734004 | 0.09 |
ENST00000467482.1
ENST00000380929.2 |
GPR143
|
G protein-coupled receptor 143 |
| chr1_-_39347255 | 0.09 |
ENST00000454994.2
ENST00000357771.3 |
GJA9
|
gap junction protein, alpha 9, 59kDa |
| chr19_-_37697976 | 0.09 |
ENST00000588873.1
|
CTC-454I21.3
|
Uncharacterized protein; Zinc finger protein 585B |
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 2.9 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 0.8 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 1.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.3 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.1 | 0.4 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.1 | 1.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.4 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.4 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.8 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 1.4 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 0.2 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 0.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 1.5 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.3 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 4.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 1.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.0 | 0.2 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.4 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.8 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.3 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.5 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.5 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.0 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.7 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0006067 | ethanol metabolic process(GO:0006067) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0071231 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.7 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 1.5 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.5 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 2.9 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 1.2 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 1.4 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 2.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 4.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 1.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.4 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 5.5 | GO:0005929 | cilium(GO:0005929) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.3 | 1.4 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.2 | 0.8 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 0.7 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 1.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 2.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.3 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.5 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 1.4 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.7 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 1.4 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 1.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.8 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.8 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 5.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 4.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.8 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 1.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |