Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
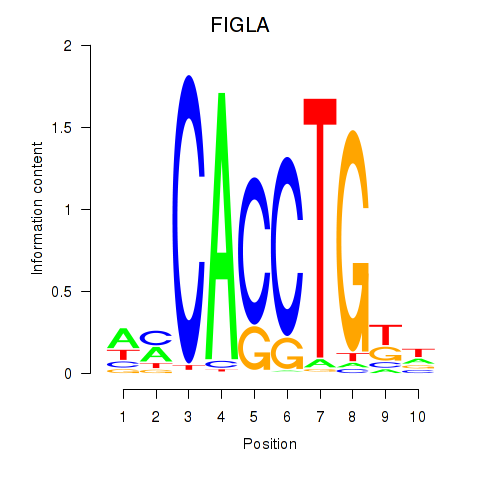
Results for FIGLA
Z-value: 0.69
Transcription factors associated with FIGLA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FIGLA
|
ENSG00000183733.6 | folliculogenesis specific bHLH transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FIGLA | hg19_v2_chr2_-_71017775_71017775 | 0.32 | 8.5e-02 | Click! |
Activity profile of FIGLA motif
Sorted Z-values of FIGLA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_39382900 | 2.90 |
ENST00000377721.3
ENST00000455970.2 |
KRTAP9-2
|
keratin associated protein 9-2 |
| chr17_-_39280419 | 1.70 |
ENST00000394014.1
|
KRTAP4-12
|
keratin associated protein 4-12 |
| chr11_+_118175596 | 1.32 |
ENST00000528600.1
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr11_+_118175132 | 1.30 |
ENST00000361763.4
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr6_-_39290316 | 1.21 |
ENST00000425054.2
ENST00000373227.4 ENST00000373229.5 ENST00000437525.2 |
KCNK16
|
potassium channel, subfamily K, member 16 |
| chr17_-_39211463 | 1.20 |
ENST00000542910.1
ENST00000398477.1 |
KRTAP2-2
|
keratin associated protein 2-2 |
| chr3_+_49059038 | 1.20 |
ENST00000451378.2
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr17_+_39411636 | 1.01 |
ENST00000394008.1
|
KRTAP9-9
|
keratin associated protein 9-9 |
| chr17_+_39261584 | 0.95 |
ENST00000391415.1
|
KRTAP4-9
|
keratin associated protein 4-9 |
| chr20_+_44441215 | 0.95 |
ENST00000356455.4
ENST00000405520.1 |
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr20_+_44441304 | 0.94 |
ENST00000352551.5
|
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr1_-_230561475 | 0.92 |
ENST00000391860.1
|
PGBD5
|
piggyBac transposable element derived 5 |
| chr17_+_39394250 | 0.78 |
ENST00000254072.6
|
KRTAP9-8
|
keratin associated protein 9-8 |
| chr2_+_42104692 | 0.78 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chr11_-_123525289 | 0.75 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr4_+_165675269 | 0.72 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr4_+_165675197 | 0.70 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr5_-_159739532 | 0.67 |
ENST00000520748.1
ENST00000393977.3 ENST00000257536.7 |
CCNJL
|
cyclin J-like |
| chr17_-_39334460 | 0.63 |
ENST00000377726.2
|
KRTAP4-2
|
keratin associated protein 4-2 |
| chr2_-_175629135 | 0.62 |
ENST00000409542.1
ENST00000409219.1 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr11_+_118958689 | 0.61 |
ENST00000535253.1
ENST00000392841.1 |
HMBS
|
hydroxymethylbilane synthase |
| chr17_+_48133459 | 0.59 |
ENST00000320031.8
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr1_-_11120057 | 0.59 |
ENST00000376957.2
|
SRM
|
spermidine synthase |
| chr7_-_99698338 | 0.59 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr11_+_60223312 | 0.58 |
ENST00000532491.1
ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr6_+_96463840 | 0.58 |
ENST00000302103.5
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase) |
| chr19_+_50180317 | 0.57 |
ENST00000534465.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr6_+_41888926 | 0.54 |
ENST00000230340.4
|
BYSL
|
bystin-like |
| chr17_-_39306054 | 0.50 |
ENST00000343246.4
|
KRTAP4-5
|
keratin associated protein 4-5 |
| chr11_+_60223225 | 0.48 |
ENST00000524807.1
ENST00000345732.4 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr19_-_51456198 | 0.48 |
ENST00000594846.1
|
KLK5
|
kallikrein-related peptidase 5 |
| chr7_+_2687173 | 0.46 |
ENST00000403167.1
|
TTYH3
|
tweety family member 3 |
| chr3_+_172468472 | 0.46 |
ENST00000232458.5
ENST00000392692.3 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr17_-_39203519 | 0.45 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr7_-_44229022 | 0.44 |
ENST00000403799.3
|
GCK
|
glucokinase (hexokinase 4) |
| chr4_-_139163491 | 0.44 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr3_-_49967292 | 0.41 |
ENST00000455683.2
|
MON1A
|
MON1 secretory trafficking family member A |
| chrX_-_30326445 | 0.41 |
ENST00000378963.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr17_+_39240459 | 0.39 |
ENST00000391417.4
|
KRTAP4-7
|
keratin associated protein 4-7 |
| chr10_-_33625154 | 0.39 |
ENST00000265371.4
|
NRP1
|
neuropilin 1 |
| chr22_-_37823468 | 0.39 |
ENST00000402918.2
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
| chr17_-_39316983 | 0.38 |
ENST00000390661.3
|
KRTAP4-4
|
keratin associated protein 4-4 |
| chr16_+_30006615 | 0.38 |
ENST00000563197.1
|
INO80E
|
INO80 complex subunit E |
| chr2_-_175629164 | 0.38 |
ENST00000409323.1
ENST00000261007.5 ENST00000348749.5 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr12_+_48357340 | 0.38 |
ENST00000256686.6
ENST00000549288.1 ENST00000552561.1 ENST00000546749.1 ENST00000552546.1 ENST00000550552.1 |
TMEM106C
|
transmembrane protein 106C |
| chr12_+_32655110 | 0.37 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr17_-_48133054 | 0.37 |
ENST00000499842.1
|
RP11-1094H24.4
|
RP11-1094H24.4 |
| chr12_-_52585765 | 0.37 |
ENST00000313234.5
ENST00000394815.2 |
KRT80
|
keratin 80 |
| chr15_+_89182178 | 0.36 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr12_+_48357401 | 0.35 |
ENST00000429772.2
ENST00000449758.2 |
TMEM106C
|
transmembrane protein 106C |
| chrX_-_137793826 | 0.34 |
ENST00000315930.6
|
FGF13
|
fibroblast growth factor 13 |
| chr17_+_8243154 | 0.32 |
ENST00000328248.2
ENST00000584943.1 |
ODF4
|
outer dense fiber of sperm tails 4 |
| chr1_+_22328144 | 0.32 |
ENST00000290122.3
ENST00000374663.1 |
CELA3A
|
chymotrypsin-like elastase family, member 3A |
| chr3_-_50360192 | 0.32 |
ENST00000442581.1
ENST00000447092.1 ENST00000357750.4 |
HYAL2
|
hyaluronoglucosaminidase 2 |
| chr1_+_183155373 | 0.31 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr19_-_42916499 | 0.30 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr12_+_58176525 | 0.30 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr15_+_90728145 | 0.30 |
ENST00000561085.1
ENST00000379122.3 ENST00000332496.6 |
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr9_+_33795533 | 0.30 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr12_-_118406777 | 0.30 |
ENST00000339824.5
|
KSR2
|
kinase suppressor of ras 2 |
| chr2_-_152590946 | 0.29 |
ENST00000172853.10
|
NEB
|
nebulin |
| chr17_+_40985407 | 0.29 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr4_-_90757364 | 0.29 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr10_-_30638090 | 0.29 |
ENST00000421701.1
ENST00000263063.4 |
MTPAP
|
mitochondrial poly(A) polymerase |
| chr7_+_18535854 | 0.29 |
ENST00000401921.1
|
HDAC9
|
histone deacetylase 9 |
| chr2_+_136289030 | 0.28 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chr20_-_22566089 | 0.28 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr2_-_163008903 | 0.28 |
ENST00000418842.2
ENST00000375497.3 |
GCG
|
glucagon |
| chr5_-_54281407 | 0.28 |
ENST00000381403.4
|
ESM1
|
endothelial cell-specific molecule 1 |
| chr11_-_84634217 | 0.28 |
ENST00000524982.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr1_+_22303503 | 0.28 |
ENST00000337107.6
|
CELA3B
|
chymotrypsin-like elastase family, member 3B |
| chr7_+_128784712 | 0.27 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr15_+_89181974 | 0.27 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr11_-_84634447 | 0.27 |
ENST00000532653.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr6_+_7107830 | 0.27 |
ENST00000379933.3
|
RREB1
|
ras responsive element binding protein 1 |
| chr19_-_40791211 | 0.27 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr6_+_44184653 | 0.26 |
ENST00000573382.2
ENST00000576476.1 |
RP1-302G2.5
|
RP1-302G2.5 |
| chr8_-_49834299 | 0.26 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr5_+_150827143 | 0.26 |
ENST00000243389.3
ENST00000517945.1 ENST00000521925.1 |
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr9_-_139258159 | 0.25 |
ENST00000371739.3
|
DNLZ
|
DNL-type zinc finger |
| chr5_+_68788594 | 0.25 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr15_+_89182156 | 0.25 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr2_+_136343820 | 0.25 |
ENST00000410054.1
|
R3HDM1
|
R3H domain containing 1 |
| chr2_+_241375069 | 0.24 |
ENST00000264039.2
|
GPC1
|
glypican 1 |
| chr8_-_49833978 | 0.24 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chrX_-_107975917 | 0.24 |
ENST00000563887.1
|
RP6-24A23.6
|
Uncharacterized protein |
| chr13_-_20767037 | 0.24 |
ENST00000382848.4
|
GJB2
|
gap junction protein, beta 2, 26kDa |
| chr2_+_170590321 | 0.24 |
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23 |
| chr1_+_22307592 | 0.23 |
ENST00000400277.2
|
CELA3B
|
chymotrypsin-like elastase family, member 3B |
| chr7_+_77428066 | 0.23 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr17_+_39405939 | 0.23 |
ENST00000334109.2
|
KRTAP9-4
|
keratin associated protein 9-4 |
| chr5_-_16936340 | 0.23 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr1_-_40367530 | 0.23 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr2_-_136288740 | 0.22 |
ENST00000264159.6
ENST00000536680.1 |
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr16_+_30006997 | 0.22 |
ENST00000304516.7
|
INO80E
|
INO80 complex subunit E |
| chr2_-_27603582 | 0.22 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr3_+_111393501 | 0.22 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr6_+_7108210 | 0.22 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr3_+_35721106 | 0.22 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr6_+_12717892 | 0.21 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr12_-_118406028 | 0.21 |
ENST00000425217.1
|
KSR2
|
kinase suppressor of ras 2 |
| chr5_-_132948216 | 0.21 |
ENST00000265342.7
|
FSTL4
|
follistatin-like 4 |
| chr6_+_34204642 | 0.21 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr4_+_6271558 | 0.21 |
ENST00000503569.1
ENST00000226760.1 |
WFS1
|
Wolfram syndrome 1 (wolframin) |
| chr1_+_26737253 | 0.21 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr6_-_31697255 | 0.21 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr11_+_128634589 | 0.21 |
ENST00000281428.8
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr17_+_7462103 | 0.21 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_+_144989309 | 0.20 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr8_-_28347737 | 0.20 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr6_+_46761118 | 0.20 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr11_-_118972575 | 0.20 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr14_-_20923195 | 0.20 |
ENST00000206542.4
|
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr7_-_37488834 | 0.20 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr8_+_32405785 | 0.20 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chr3_-_194991876 | 0.20 |
ENST00000310380.6
|
XXYLT1
|
xyloside xylosyltransferase 1 |
| chr5_+_6448736 | 0.20 |
ENST00000399816.3
|
UBE2QL1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr16_-_69418649 | 0.20 |
ENST00000566257.1
|
TERF2
|
telomeric repeat binding factor 2 |
| chr2_+_37571845 | 0.19 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr16_-_46864955 | 0.19 |
ENST00000565112.1
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr1_-_201391149 | 0.19 |
ENST00000555948.1
ENST00000556362.1 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr22_+_22988816 | 0.19 |
ENST00000480559.1
ENST00000448514.1 |
GGTLC2
|
gamma-glutamyltransferase light chain 2 |
| chr6_+_22569784 | 0.19 |
ENST00000510882.2
|
HDGFL1
|
hepatoma derived growth factor-like 1 |
| chr11_+_69455855 | 0.19 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr8_+_145438870 | 0.19 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B |
| chr1_-_183622442 | 0.19 |
ENST00000308641.4
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr17_+_7461849 | 0.18 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_-_171621815 | 0.18 |
ENST00000037502.6
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response |
| chr2_+_37571717 | 0.18 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr11_-_88070920 | 0.18 |
ENST00000524463.1
ENST00000227266.5 |
CTSC
|
cathepsin C |
| chr2_-_152590982 | 0.18 |
ENST00000409198.1
ENST00000397345.3 ENST00000427231.2 |
NEB
|
nebulin |
| chr11_-_62911693 | 0.18 |
ENST00000417740.1
ENST00000326192.5 |
SLC22A24
|
solute carrier family 22, member 24 |
| chr7_-_11871815 | 0.17 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr11_-_118122996 | 0.17 |
ENST00000525386.1
ENST00000527472.1 ENST00000278949.4 |
MPZL3
|
myelin protein zero-like 3 |
| chr8_-_93029520 | 0.17 |
ENST00000521553.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr10_+_115614370 | 0.17 |
ENST00000369301.3
|
NHLRC2
|
NHL repeat containing 2 |
| chrX_-_134049233 | 0.17 |
ENST00000370779.4
|
MOSPD1
|
motile sperm domain containing 1 |
| chr2_-_128051670 | 0.16 |
ENST00000493187.2
|
ERCC3
|
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
| chr11_+_130318869 | 0.16 |
ENST00000299164.2
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15 |
| chr14_+_75746340 | 0.16 |
ENST00000555686.1
ENST00000555672.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr1_-_42384343 | 0.16 |
ENST00000372584.1
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr22_+_31090793 | 0.16 |
ENST00000332585.6
ENST00000382310.3 ENST00000446658.2 |
OSBP2
|
oxysterol binding protein 2 |
| chr7_-_148725733 | 0.16 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr5_+_140868717 | 0.16 |
ENST00000252087.1
|
PCDHGC5
|
protocadherin gamma subfamily C, 5 |
| chr1_-_156675535 | 0.15 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr1_-_111991850 | 0.15 |
ENST00000411751.2
|
WDR77
|
WD repeat domain 77 |
| chr1_+_24646002 | 0.15 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr5_+_52856456 | 0.15 |
ENST00000296684.5
ENST00000506765.1 |
NDUFS4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) |
| chr1_+_24645807 | 0.15 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_+_24645865 | 0.15 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr17_-_4458616 | 0.15 |
ENST00000381556.2
|
MYBBP1A
|
MYB binding protein (P160) 1a |
| chr1_-_53608289 | 0.14 |
ENST00000371491.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr3_-_128294929 | 0.14 |
ENST00000356020.2
|
C3orf27
|
chromosome 3 open reading frame 27 |
| chr19_+_36347787 | 0.14 |
ENST00000347900.6
ENST00000360202.5 |
KIRREL2
|
kin of IRRE like 2 (Drosophila) |
| chr19_+_36249044 | 0.14 |
ENST00000444637.2
ENST00000396908.4 ENST00000544099.1 |
C19orf55
|
chromosome 19 open reading frame 55 |
| chr16_-_4465886 | 0.14 |
ENST00000539968.1
|
CORO7
|
coronin 7 |
| chr8_-_130951940 | 0.14 |
ENST00000522250.1
ENST00000522941.1 ENST00000522746.1 ENST00000520204.1 ENST00000519070.1 ENST00000520254.1 ENST00000519824.2 ENST00000519540.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr12_+_79258444 | 0.14 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr8_-_145018905 | 0.14 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr2_-_179672142 | 0.14 |
ENST00000342992.6
ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN
|
titin |
| chr2_-_50574856 | 0.14 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr14_+_75746781 | 0.14 |
ENST00000555347.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr2_-_75788038 | 0.14 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr17_+_7462031 | 0.13 |
ENST00000380535.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_-_28969517 | 0.13 |
ENST00000263974.4
ENST00000373824.4 |
TAF12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
| chr17_+_9548845 | 0.13 |
ENST00000570475.1
ENST00000285199.7 |
USP43
|
ubiquitin specific peptidase 43 |
| chr4_-_57522470 | 0.13 |
ENST00000503639.3
|
HOPX
|
HOP homeobox |
| chr3_-_46904946 | 0.13 |
ENST00000292327.4
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr17_+_79495397 | 0.13 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr17_-_34890665 | 0.12 |
ENST00000586007.1
|
MYO19
|
myosin XIX |
| chr3_+_68055366 | 0.12 |
ENST00000496687.1
|
FAM19A1
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
| chr16_+_6533380 | 0.12 |
ENST00000552089.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr17_-_39222131 | 0.12 |
ENST00000394015.2
|
KRTAP2-4
|
keratin associated protein 2-4 |
| chr3_-_46904918 | 0.12 |
ENST00000395869.1
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr14_+_31343747 | 0.12 |
ENST00000216361.4
ENST00000396618.3 ENST00000475087.1 |
COCH
|
cochlin |
| chr12_+_79258547 | 0.12 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr6_+_109169591 | 0.12 |
ENST00000368972.3
ENST00000392644.4 |
ARMC2
|
armadillo repeat containing 2 |
| chr1_-_156675368 | 0.12 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr19_+_16186903 | 0.12 |
ENST00000588507.1
|
TPM4
|
tropomyosin 4 |
| chrX_-_134049262 | 0.12 |
ENST00000370783.3
|
MOSPD1
|
motile sperm domain containing 1 |
| chr6_-_35888824 | 0.12 |
ENST00000361690.3
ENST00000512445.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr7_+_18535346 | 0.11 |
ENST00000405010.3
ENST00000406451.4 ENST00000428307.2 |
HDAC9
|
histone deacetylase 9 |
| chr3_-_167191814 | 0.11 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr11_-_65629497 | 0.11 |
ENST00000532134.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr16_+_32077386 | 0.11 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chrX_-_11445856 | 0.11 |
ENST00000380736.1
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chrX_+_70443050 | 0.11 |
ENST00000361726.6
|
GJB1
|
gap junction protein, beta 1, 32kDa |
| chr3_-_69435224 | 0.11 |
ENST00000398540.3
|
FRMD4B
|
FERM domain containing 4B |
| chr17_-_39254391 | 0.11 |
ENST00000333822.4
|
KRTAP4-8
|
keratin associated protein 4-8 |
| chr1_+_155051305 | 0.11 |
ENST00000368408.3
|
EFNA3
|
ephrin-A3 |
| chr16_+_30383613 | 0.11 |
ENST00000568749.1
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr12_+_69201923 | 0.11 |
ENST00000462284.1
ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr14_-_107131560 | 0.11 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr1_-_153585539 | 0.11 |
ENST00000368706.4
|
S100A16
|
S100 calcium binding protein A16 |
| chr1_+_45478568 | 0.10 |
ENST00000428106.1
|
UROD
|
uroporphyrinogen decarboxylase |
| chr6_-_19804973 | 0.10 |
ENST00000457670.1
ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2
|
RP4-625H18.2 |
| chr6_-_31938700 | 0.10 |
ENST00000495340.1
|
DXO
|
decapping exoribonuclease |
| chr17_-_29624343 | 0.10 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr7_-_2883928 | 0.10 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr14_+_62584197 | 0.10 |
ENST00000334389.4
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FIGLA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 | 1.9 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.2 | 0.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 0.5 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.6 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.7 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.4 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 1.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.3 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.7 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.3 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.1 | 0.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.6 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.6 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.5 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.3 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.1 | 0.4 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 0.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.2 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.3 | GO:0009644 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.1 | 0.2 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.2 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.1 | 0.3 | GO:2000470 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.4 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 0.3 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.2 | GO:0030167 | proteoglycan catabolic process(GO:0030167) heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.5 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.6 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.3 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.3 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.6 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 7.0 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.5 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.3 | GO:0090050 | peptidyl-lysine deacetylation(GO:0034983) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.6 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.7 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.6 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.3 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.3 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 1.2 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0032425 | positive regulation of mismatch repair(GO:0032425) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0051511 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.2 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 0.6 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.1 | 8.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.3 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.1 | 0.5 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.3 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 1.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.2 | 2.6 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.2 | 0.6 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 0.4 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.7 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.4 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.3 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 0.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.6 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.3 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.6 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.4 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.3 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 0.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 1.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 0.2 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 2.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.1 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.2 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.4 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |