Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
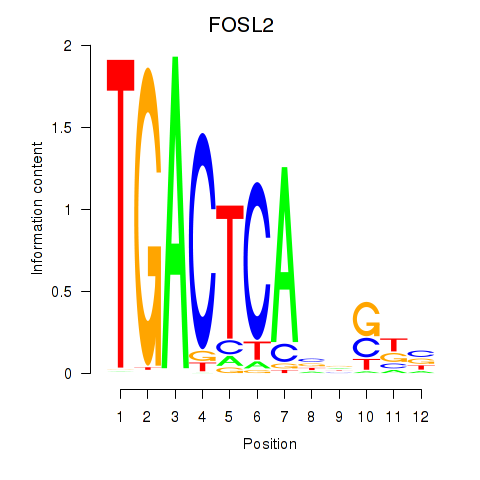
Results for FOSL2_SMARCC1
Z-value: 2.39
Transcription factors associated with FOSL2_SMARCC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSL2
|
ENSG00000075426.7 | FOS like 2, AP-1 transcription factor subunit |
|
SMARCC1
|
ENSG00000173473.6 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOSL2 | hg19_v2_chr2_+_28618532_28618610 | 0.86 | 1.7e-09 | Click! |
| SMARCC1 | hg19_v2_chr3_-_47823298_47823423 | -0.42 | 2.0e-02 | Click! |
Activity profile of FOSL2_SMARCC1 motif
Sorted Z-values of FOSL2_SMARCC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_21452964 | 34.22 |
ENST00000587184.1
|
LAMA3
|
laminin, alpha 3 |
| chr18_+_21452804 | 29.68 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr1_+_150480551 | 27.54 |
ENST00000369049.4
ENST00000369047.4 |
ECM1
|
extracellular matrix protein 1 |
| chr1_+_150480576 | 27.23 |
ENST00000346569.6
|
ECM1
|
extracellular matrix protein 1 |
| chr19_-_36019123 | 25.28 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr17_-_39769005 | 19.21 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr1_-_153521597 | 15.86 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr2_+_113875466 | 14.55 |
ENST00000361779.3
ENST00000259206.5 ENST00000354115.2 |
IL1RN
|
interleukin 1 receptor antagonist |
| chr1_-_153029980 | 14.17 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr6_+_74405804 | 12.94 |
ENST00000287097.5
|
CD109
|
CD109 molecule |
| chr1_-_153521714 | 12.88 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr6_+_74405501 | 12.79 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr11_-_65667997 | 12.72 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chr1_-_153113927 | 12.27 |
ENST00000368752.4
|
SPRR2B
|
small proline-rich protein 2B |
| chr19_-_51472823 | 11.95 |
ENST00000310157.2
|
KLK6
|
kallikrein-related peptidase 6 |
| chr15_-_74504560 | 11.70 |
ENST00000449139.2
|
STRA6
|
stimulated by retinoic acid 6 |
| chr11_-_65667884 | 10.82 |
ENST00000448083.2
ENST00000531493.1 ENST00000532401.1 |
FOSL1
|
FOS-like antigen 1 |
| chr12_-_95510743 | 9.78 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr6_+_47666275 | 9.59 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr11_-_62323702 | 9.56 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr18_+_34124507 | 9.49 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr12_+_13349650 | 9.46 |
ENST00000256951.5
ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1
|
epithelial membrane protein 1 |
| chr18_+_61442629 | 9.22 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr19_-_51504411 | 9.19 |
ENST00000593490.1
|
KLK8
|
kallikrein-related peptidase 8 |
| chr3_-_48632593 | 9.17 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr11_-_102668879 | 9.16 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr5_-_176923846 | 8.96 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr1_-_205419053 | 8.91 |
ENST00000367154.1
|
LEMD1
|
LEM domain containing 1 |
| chr12_-_48152853 | 8.55 |
ENST00000171000.4
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr12_-_95009837 | 8.37 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr9_+_140119618 | 8.33 |
ENST00000359069.2
|
C9orf169
|
chromosome 9 open reading frame 169 |
| chr12_-_48152611 | 8.19 |
ENST00000389212.3
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr17_+_4853442 | 8.00 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr10_+_17270214 | 7.96 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr11_-_6341844 | 7.88 |
ENST00000303927.3
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr5_-_176923803 | 7.82 |
ENST00000506161.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr7_+_48128194 | 7.81 |
ENST00000416681.1
ENST00000331803.4 ENST00000432131.1 |
UPP1
|
uridine phosphorylase 1 |
| chr7_+_48128316 | 7.79 |
ENST00000341253.4
|
UPP1
|
uridine phosphorylase 1 |
| chr5_+_135394840 | 7.11 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr13_+_78109884 | 7.01 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr19_-_51538118 | 6.92 |
ENST00000529888.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr19_-_35992780 | 6.91 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr11_+_12308447 | 6.89 |
ENST00000256186.2
|
MICALCL
|
MICAL C-terminal like |
| chr20_-_634000 | 6.82 |
ENST00000381962.3
|
SRXN1
|
sulfiredoxin 1 |
| chr13_+_78109804 | 6.77 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr19_-_51538148 | 6.65 |
ENST00000319590.4
ENST00000250351.4 |
KLK12
|
kallikrein-related peptidase 12 |
| chr11_-_66103932 | 6.41 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr2_-_113594279 | 6.33 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr19_-_36004543 | 6.32 |
ENST00000339686.3
ENST00000447113.2 ENST00000440396.1 |
DMKN
|
dermokine |
| chr1_+_183155373 | 6.25 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr19_-_51523412 | 6.09 |
ENST00000391805.1
ENST00000599077.1 |
KLK10
|
kallikrein-related peptidase 10 |
| chr20_+_33759854 | 6.00 |
ENST00000216968.4
|
PROCR
|
protein C receptor, endothelial |
| chr19_+_39279838 | 5.98 |
ENST00000314980.4
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B |
| chr2_-_113542063 | 5.93 |
ENST00000263339.3
|
IL1A
|
interleukin 1, alpha |
| chr5_+_149877334 | 5.92 |
ENST00000523767.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr12_-_122238464 | 5.84 |
ENST00000546227.1
|
RHOF
|
ras homolog family member F (in filopodia) |
| chr1_+_154377669 | 5.77 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr15_-_74504597 | 5.76 |
ENST00000416286.3
|
STRA6
|
stimulated by retinoic acid 6 |
| chr11_-_66104237 | 5.67 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr11_-_66103867 | 5.66 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr13_+_32838801 | 5.63 |
ENST00000542859.1
|
FRY
|
furry homolog (Drosophila) |
| chr12_+_53491220 | 5.48 |
ENST00000548547.1
ENST00000301464.3 |
IGFBP6
|
insulin-like growth factor binding protein 6 |
| chr15_+_67420441 | 5.40 |
ENST00000558894.1
|
SMAD3
|
SMAD family member 3 |
| chr9_+_35673853 | 5.36 |
ENST00000378357.4
|
CA9
|
carbonic anhydrase IX |
| chrX_+_135251783 | 5.08 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr19_-_44174330 | 5.07 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr15_-_80263506 | 5.06 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr17_+_74381343 | 4.87 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr4_-_39033963 | 4.86 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr18_+_61445007 | 4.82 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr1_-_28520384 | 4.78 |
ENST00000305392.3
|
PTAFR
|
platelet-activating factor receptor |
| chr19_-_51523275 | 4.73 |
ENST00000309958.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr12_-_52845910 | 4.71 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chrX_+_135252050 | 4.70 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr7_-_107643674 | 4.68 |
ENST00000222399.6
|
LAMB1
|
laminin, beta 1 |
| chr16_+_58533951 | 4.65 |
ENST00000566192.1
ENST00000565088.1 ENST00000568640.1 ENST00000563978.1 ENST00000569923.1 ENST00000356752.4 ENST00000563799.1 ENST00000562999.1 ENST00000570248.1 ENST00000562731.1 ENST00000568424.1 |
NDRG4
|
NDRG family member 4 |
| chrX_+_135251835 | 4.63 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chrX_+_99899180 | 4.61 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr6_+_41604747 | 4.56 |
ENST00000419164.1
ENST00000373051.2 |
MDFI
|
MyoD family inhibitor |
| chr1_+_26606608 | 4.54 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr15_+_67458357 | 4.53 |
ENST00000537194.2
|
SMAD3
|
SMAD family member 3 |
| chr12_-_49259643 | 4.52 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr3_-_98241358 | 4.52 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr10_-_90611566 | 4.46 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr1_-_28520447 | 4.41 |
ENST00000539896.1
|
PTAFR
|
platelet-activating factor receptor |
| chr17_-_7493390 | 4.39 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr19_-_44174305 | 4.37 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr1_-_204121013 | 4.34 |
ENST00000367201.3
|
ETNK2
|
ethanolamine kinase 2 |
| chr4_+_74606223 | 4.32 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr1_-_204121102 | 4.31 |
ENST00000367202.4
|
ETNK2
|
ethanolamine kinase 2 |
| chr20_+_44637526 | 4.31 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr16_-_30125177 | 4.25 |
ENST00000406256.3
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr1_-_204121298 | 4.21 |
ENST00000367199.2
|
ETNK2
|
ethanolamine kinase 2 |
| chr1_-_156675368 | 4.08 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr19_-_43702231 | 4.07 |
ENST00000597374.1
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr17_-_27503770 | 4.05 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr11_-_6341724 | 4.01 |
ENST00000530979.1
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr19_-_44285401 | 3.96 |
ENST00000262888.3
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr18_+_56338750 | 3.95 |
ENST00000345724.3
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chr11_+_57308979 | 3.92 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr17_+_7482785 | 3.90 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr8_+_87111059 | 3.90 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr20_+_36012051 | 3.86 |
ENST00000373567.2
|
SRC
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr6_+_32121218 | 3.83 |
ENST00000414204.1
ENST00000361568.2 ENST00000395523.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr11_+_62649158 | 3.81 |
ENST00000539891.1
ENST00000536981.1 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr19_-_51568324 | 3.81 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr9_-_35112376 | 3.77 |
ENST00000488109.2
|
FAM214B
|
family with sequence similarity 214, member B |
| chr11_+_394196 | 3.74 |
ENST00000331563.2
ENST00000531857.1 |
PKP3
|
plakophilin 3 |
| chr11_+_844406 | 3.72 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr15_+_67418047 | 3.69 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr4_-_39034542 | 3.68 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr1_-_159924006 | 3.65 |
ENST00000368092.3
ENST00000368093.3 |
SLAMF9
|
SLAM family member 9 |
| chr1_-_150208320 | 3.61 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr6_+_106546808 | 3.55 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr3_-_12800751 | 3.55 |
ENST00000435218.2
ENST00000435575.1 |
TMEM40
|
transmembrane protein 40 |
| chr2_+_90060377 | 3.55 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr4_-_175443943 | 3.43 |
ENST00000296522.6
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr1_-_179112173 | 3.39 |
ENST00000408940.3
ENST00000504405.1 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr16_-_122619 | 3.38 |
ENST00000262316.6
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila) |
| chr17_+_73717407 | 3.36 |
ENST00000579662.1
|
ITGB4
|
integrin, beta 4 |
| chr2_+_89998789 | 3.33 |
ENST00000453166.2
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr2_+_87755054 | 3.31 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr17_-_41623075 | 3.30 |
ENST00000545089.1
|
ETV4
|
ets variant 4 |
| chr1_+_26605618 | 3.29 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr2_+_87754989 | 3.28 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr10_+_75670862 | 3.25 |
ENST00000446342.1
ENST00000372764.3 ENST00000372762.4 |
PLAU
|
plasminogen activator, urokinase |
| chr19_-_39264072 | 3.25 |
ENST00000599035.1
ENST00000378626.4 |
LGALS7
|
lectin, galactoside-binding, soluble, 7 |
| chr1_-_151965048 | 3.23 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10 |
| chr19_-_6670128 | 3.22 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr6_-_35888824 | 3.21 |
ENST00000361690.3
ENST00000512445.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr22_-_32555275 | 3.19 |
ENST00000382097.3
|
C22orf42
|
chromosome 22 open reading frame 42 |
| chr3_-_47950745 | 3.17 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr8_+_10530133 | 3.17 |
ENST00000304519.5
|
C8orf74
|
chromosome 8 open reading frame 74 |
| chr2_+_87754887 | 3.15 |
ENST00000409054.1
ENST00000331944.6 ENST00000409139.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr11_+_35639735 | 3.14 |
ENST00000317811.4
|
FJX1
|
four jointed box 1 (Drosophila) |
| chr2_-_89521942 | 3.14 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr6_-_131277510 | 3.11 |
ENST00000525193.1
ENST00000527659.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr9_-_35685452 | 3.07 |
ENST00000607559.1
|
TPM2
|
tropomyosin 2 (beta) |
| chr1_+_156095951 | 3.00 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr11_+_35198243 | 3.00 |
ENST00000528455.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr18_+_61254570 | 2.97 |
ENST00000344731.5
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr2_+_89999259 | 2.95 |
ENST00000558026.1
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr6_-_35888905 | 2.89 |
ENST00000510290.1
ENST00000423325.2 ENST00000373822.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr1_+_201252580 | 2.89 |
ENST00000367324.3
ENST00000263946.3 |
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) |
| chr2_-_85641162 | 2.87 |
ENST00000447219.2
ENST00000409670.1 ENST00000409724.1 |
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr2_-_89459813 | 2.87 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr9_-_130341268 | 2.86 |
ENST00000373314.3
|
FAM129B
|
family with sequence similarity 129, member B |
| chrX_-_15683147 | 2.85 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr7_+_129932974 | 2.85 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr3_-_81792780 | 2.84 |
ENST00000489715.1
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr11_-_118135160 | 2.84 |
ENST00000438295.2
|
MPZL2
|
myelin protein zero-like 2 |
| chr11_-_82708519 | 2.81 |
ENST00000534301.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr3_-_149095652 | 2.80 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr17_+_73717551 | 2.80 |
ENST00000450894.3
|
ITGB4
|
integrin, beta 4 |
| chr7_+_134528635 | 2.75 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr3_+_154797877 | 2.74 |
ENST00000462745.1
ENST00000493237.1 |
MME
|
membrane metallo-endopeptidase |
| chr12_-_6960407 | 2.70 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr18_+_56338618 | 2.68 |
ENST00000348428.3
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chr18_+_52495426 | 2.65 |
ENST00000262094.5
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr14_-_91710852 | 2.64 |
ENST00000535815.1
ENST00000529102.1 |
GPR68
|
G protein-coupled receptor 68 |
| chr2_+_27193480 | 2.63 |
ENST00000233121.2
ENST00000405074.3 |
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr20_-_1309809 | 2.62 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr2_+_191792376 | 2.60 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr5_-_176924562 | 2.60 |
ENST00000359895.2
ENST00000355572.2 ENST00000355841.2 ENST00000393551.1 ENST00000505074.1 ENST00000356618.4 ENST00000393546.4 |
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr11_+_35198118 | 2.59 |
ENST00000525211.1
ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr3_+_11178779 | 2.57 |
ENST00000438284.2
|
HRH1
|
histamine receptor H1 |
| chr7_+_55177416 | 2.54 |
ENST00000450046.1
ENST00000454757.2 |
EGFR
|
epidermal growth factor receptor |
| chr19_+_926000 | 2.53 |
ENST00000263620.3
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr14_+_96722539 | 2.53 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chrX_+_49028265 | 2.50 |
ENST00000376322.3
ENST00000376327.5 |
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched) |
| chr17_+_79650962 | 2.48 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr10_-_4285923 | 2.47 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr16_+_30006615 | 2.44 |
ENST00000563197.1
|
INO80E
|
INO80 complex subunit E |
| chr4_-_987217 | 2.44 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr14_+_96722152 | 2.43 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr22_+_21987005 | 2.42 |
ENST00000607942.1
ENST00000425975.1 ENST00000292779.3 |
CCDC116
|
coiled-coil domain containing 116 |
| chr14_+_35591858 | 2.42 |
ENST00000603544.1
|
KIAA0391
|
KIAA0391 |
| chrX_+_47441712 | 2.42 |
ENST00000218388.4
ENST00000377018.2 ENST00000456754.2 ENST00000377017.1 ENST00000441738.1 |
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr2_+_220492373 | 2.41 |
ENST00000317151.3
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr2_+_169926047 | 2.37 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr17_-_41623259 | 2.36 |
ENST00000538265.1
ENST00000591713.1 |
ETV4
|
ets variant 4 |
| chr1_-_150208291 | 2.36 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr3_-_87040233 | 2.35 |
ENST00000398399.2
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr1_-_150208363 | 2.33 |
ENST00000436748.2
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr11_-_64013663 | 2.33 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr6_-_30712313 | 2.33 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr1_-_154943212 | 2.32 |
ENST00000368445.5
ENST00000448116.2 ENST00000368449.4 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr3_+_5020801 | 2.32 |
ENST00000256495.3
|
BHLHE40
|
basic helix-loop-helix family, member e40 |
| chr12_+_53440753 | 2.32 |
ENST00000379902.3
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr5_-_141061777 | 2.30 |
ENST00000239440.4
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr1_+_901847 | 2.29 |
ENST00000379410.3
ENST00000379409.2 ENST00000379407.3 |
PLEKHN1
|
pleckstrin homology domain containing, family N member 1 |
| chr17_-_33390667 | 2.28 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr19_+_17865011 | 2.25 |
ENST00000596462.1
ENST00000596865.1 ENST00000598960.1 ENST00000539407.1 |
FCHO1
|
FCH domain only 1 |
| chr18_+_268148 | 2.25 |
ENST00000581677.1
|
RP11-705O1.8
|
RP11-705O1.8 |
| chr3_+_52350335 | 2.25 |
ENST00000420323.2
|
DNAH1
|
dynein, axonemal, heavy chain 1 |
| chr12_-_57914275 | 2.24 |
ENST00000547303.1
ENST00000552740.1 ENST00000547526.1 ENST00000551116.1 ENST00000346473.3 |
DDIT3
|
DNA-damage-inducible transcript 3 |
| chr1_-_21059029 | 2.20 |
ENST00000444387.2
ENST00000375031.1 ENST00000518294.1 |
SH2D5
|
SH2 domain containing 5 |
| chr1_-_154943002 | 2.18 |
ENST00000606391.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr5_-_141061759 | 2.16 |
ENST00000508305.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr15_-_56209306 | 2.14 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOSL2_SMARCC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 16.7 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 3.9 | 23.5 | GO:0007296 | vitellogenesis(GO:0007296) |
| 3.9 | 15.6 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 3.8 | 22.9 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 3.5 | 14.0 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 3.5 | 17.5 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 3.1 | 9.2 | GO:1904300 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 2.8 | 82.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 2.7 | 13.6 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 2.4 | 9.4 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 2.2 | 17.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 2.2 | 13.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 2.1 | 6.3 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 2.1 | 54.7 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 1.7 | 5.2 | GO:0002384 | hepatic immune response(GO:0002384) |
| 1.6 | 4.9 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 1.5 | 4.4 | GO:0048627 | myoblast development(GO:0048627) |
| 1.4 | 11.0 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 1.3 | 4.0 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 1.3 | 6.6 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 1.3 | 9.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 1.3 | 23.5 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 1.2 | 4.7 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 1.1 | 3.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 1.0 | 4.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 1.0 | 3.9 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 1.0 | 2.9 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.9 | 4.5 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.8 | 5.9 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.8 | 4.1 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.8 | 5.6 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.8 | 2.3 | GO:0014806 | negative regulation of muscle hyperplasia(GO:0014740) smooth muscle hyperplasia(GO:0014806) |
| 0.8 | 3.8 | GO:0060356 | leucine import(GO:0060356) |
| 0.7 | 3.0 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.7 | 2.9 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.7 | 3.6 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.7 | 5.6 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.7 | 3.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.7 | 2.1 | GO:0045659 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.7 | 8.2 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.6 | 3.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.6 | 4.5 | GO:1902722 | positive regulation of prolactin secretion(GO:1902722) |
| 0.6 | 1.9 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.6 | 0.6 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.6 | 3.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.6 | 1.8 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.6 | 1.8 | GO:2000452 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) positive regulation of necroptotic process(GO:0060545) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.6 | 9.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.6 | 5.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.6 | 4.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.6 | 2.8 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.5 | 1.6 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.5 | 3.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.5 | 2.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.5 | 8.8 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.5 | 0.5 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.5 | 3.4 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.5 | 12.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.5 | 1.4 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.5 | 4.6 | GO:0032962 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.5 | 2.3 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 | 2.2 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) regulation of determination of dorsal identity(GO:2000015) |
| 0.4 | 3.6 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.4 | 6.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.4 | 1.3 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.4 | 4.7 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.4 | 3.8 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.4 | 4.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.4 | 1.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.4 | 4.1 | GO:0042325 | regulation of phosphorylation(GO:0042325) |
| 0.4 | 2.9 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.4 | 1.6 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.4 | 1.6 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.4 | 1.2 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.4 | 3.8 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.4 | 1.5 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.4 | 1.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.4 | 1.9 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.4 | 1.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.4 | 9.5 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 2.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.4 | 2.9 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.4 | 1.1 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.4 | 5.3 | GO:0044351 | macropinocytosis(GO:0044351) phagosome-lysosome fusion(GO:0090385) |
| 0.3 | 1.0 | GO:0060621 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.3 | 4.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 24.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.3 | 2.6 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.3 | 1.0 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.3 | 1.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 | 1.0 | GO:0071451 | cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.3 | 0.9 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.3 | 2.5 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.3 | 4.7 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.3 | 4.9 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.3 | 4.6 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.3 | 2.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.3 | 0.9 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.3 | 0.8 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.3 | 1.9 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.3 | 0.8 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.3 | 0.8 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.3 | 1.9 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.3 | 5.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.3 | 23.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.3 | 12.2 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.3 | 1.0 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.3 | 1.3 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.2 | 0.2 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.2 | 1.2 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.2 | 1.0 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.2 | 3.2 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.2 | 3.0 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.2 | 1.7 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.2 | 11.7 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 1.2 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.2 | 0.7 | GO:0032639 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.2 | 1.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 5.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 2.4 | GO:1905049 | negative regulation of metallopeptidase activity(GO:1905049) |
| 0.2 | 3.5 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.2 | 0.7 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.2 | 0.9 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.2 | 2.6 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.2 | 0.6 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 13.2 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.2 | 0.6 | GO:0050894 | determination of affect(GO:0050894) |
| 0.2 | 3.1 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 2.9 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 0.6 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.2 | 1.2 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 1.2 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.2 | 1.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 4.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 2.6 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.2 | 2.0 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.2 | 1.7 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.2 | 0.6 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.2 | 17.8 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.2 | 0.6 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.2 | 1.3 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.2 | 1.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 1.0 | GO:0030421 | defecation(GO:0030421) |
| 0.2 | 4.7 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.2 | 2.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.2 | 1.0 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.2 | 2.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 0.6 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 5.0 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 3.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.2 | 2.5 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.2 | 1.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.2 | 2.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 2.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 2.3 | GO:2001020 | regulation of response to DNA damage stimulus(GO:2001020) |
| 0.1 | 0.3 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.1 | 0.8 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 0.1 | 7.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 2.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.5 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 9.4 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.1 | 6.4 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.1 | 0.6 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.5 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 3.9 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 0.7 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.1 | 0.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 1.0 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 4.4 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 2.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.6 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) Schwann cell proliferation(GO:0014010) |
| 0.1 | 1.4 | GO:0050777 | negative regulation of immune response(GO:0050777) |
| 0.1 | 3.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 10.1 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 2.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 5.9 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 2.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 1.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.9 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 1.6 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.5 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 1.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 4.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 1.6 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.1 | 0.5 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 0.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.7 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) negative regulation of amino acid transport(GO:0051956) |
| 0.1 | 1.2 | GO:0035588 | G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.5 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.7 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.9 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 3.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.2 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.1 | GO:0014040 | positive regulation of Schwann cell differentiation(GO:0014040) |
| 0.1 | 1.8 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 1.6 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.1 | 0.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 3.8 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.5 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 1.2 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 1.3 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.1 | 0.9 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.8 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.2 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.1 | 1.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.8 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.9 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.1 | 0.2 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.1 | 1.8 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 1.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.2 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 3.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.3 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.7 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 3.1 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 2.5 | GO:0035556 | intracellular signal transduction(GO:0035556) |
| 0.0 | 0.3 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 2.0 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 1.5 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.3 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 1.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 1.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 2.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.6 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.7 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.8 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 2.5 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 | 1.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 5.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 1.0 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.6 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.9 | GO:0006520 | cellular amino acid metabolic process(GO:0006520) |
| 0.0 | 4.0 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 1.2 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 1.2 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.2 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.6 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 2.5 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.9 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) optic nerve structural organization(GO:0021633) |
| 0.0 | 0.4 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 1.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.4 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 1.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.7 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 2.5 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 1.7 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 1.5 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.6 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.0 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.4 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.9 | GO:0003231 | cardiac ventricle development(GO:0003231) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 2.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 1.2 | GO:0048207 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.7 | GO:0006090 | pyruvate metabolic process(GO:0006090) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 1.7 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 1.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 2.3 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.0 | GO:0035787 | cell migration involved in kidney development(GO:0035787) cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) lung growth(GO:0060437) angiogenesis involved in coronary vascular morphogenesis(GO:0060978) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.1 | 65.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 3.6 | 10.9 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 3.0 | 54.7 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 2.3 | 9.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 1.7 | 13.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 1.5 | 10.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.3 | 6.6 | GO:0032449 | CBM complex(GO:0032449) |
| 1.3 | 5.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 1.2 | 7.0 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 1.0 | 11.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.9 | 3.7 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.8 | 2.5 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.8 | 28.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.8 | 5.6 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.7 | 2.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.7 | 44.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.6 | 3.8 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.6 | 2.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.6 | 11.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.5 | 5.5 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.5 | 4.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 1.9 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.5 | 1.9 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.4 | 9.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 1.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 4.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 3.9 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.3 | 10.4 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.0 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.2 | 5.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.2 | 31.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 2.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 1.7 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 23.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 2.9 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 1.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.2 | 9.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 2.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 2.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 22.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 0.7 | GO:0019031 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.2 | 1.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 2.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 4.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 41.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 1.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 1.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 3.4 | GO:0097346 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) INO80-type complex(GO:0097346) |
| 0.1 | 1.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 3.2 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 4.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.8 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.6 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 2.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 12.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 0.5 | GO:0002079 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
| 0.1 | 1.0 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 1.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.4 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 4.1 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 23.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 3.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 3.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.8 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 0.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 1.7 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 2.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 5.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.6 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 2.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 5.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.9 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.7 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.2 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 4.6 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 1.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.8 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.1 | 0.3 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.7 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 4.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 12.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 6.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 4.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 3.8 | GO:0031253 | cell projection membrane(GO:0031253) |
| 0.0 | 44.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 15.6 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.5 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.0 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 1.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 39.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.3 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.8 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.5 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.3 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 4.5 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0032155 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.0 | 1.7 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.2 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.8 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 0.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 54.8 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 4.9 | 14.6 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 3.1 | 9.4 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 3.1 | 15.6 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 2.7 | 13.6 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 1.9 | 5.8 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 1.8 | 9.2 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 1.7 | 5.0 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 1.5 | 10.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.5 | 5.9 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 1.4 | 16.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 1.3 | 9.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.3 | 12.9 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 1.2 | 3.5 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.0 | 7.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.9 | 3.4 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.8 | 4.9 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.7 | 13.4 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.7 | 26.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.6 | 3.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.6 | 1.9 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.6 | 2.5 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.6 | 12.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.6 | 1.7 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.6 | 2.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.6 | 1.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.6 | 2.8 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.5 | 2.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.5 | 1.9 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.5 | 1.4 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.5 | 1.4 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.4 | 4.7 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.4 | 2.6 | GO:0042806 | fucose binding(GO:0042806) |
| 0.4 | 3.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.4 | 2.9 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.4 | 8.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 14.8 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.4 | 3.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.4 | 3.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.4 | 1.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.4 | 3.8 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.4 | 5.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.4 | 1.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.4 | 4.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.4 | 1.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 2.1 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 5.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 9.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.3 | 1.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.3 | 1.3 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.3 | 2.0 | GO:0042835 | BRE binding(GO:0042835) |
| 0.3 | 1.3 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.3 | 0.9 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.3 | 3.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.3 | 5.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 1.2 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.3 | 1.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 1.6 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.3 | 0.8 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.3 | 2.1 | GO:0050815 | phosphoserine binding(GO:0050815) phosphothreonine binding(GO:0050816) |
| 0.3 | 1.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.3 | 2.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 1.0 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.2 | 1.0 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 3.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 3.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 2.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 2.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 5.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 15.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 0.7 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 0.6 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.2 | 66.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 6.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 0.6 | GO:0031711 | angiotensin type I receptor activity(GO:0001596) bradykinin receptor binding(GO:0031711) |
| 0.2 | 25.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.6 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.2 | 4.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 0.6 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 6.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 0.9 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.2 | 4.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 1.9 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.2 | 1.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 0.6 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.2 | 2.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 1.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 6.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 2.1 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.4 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 1.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 4.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 10.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 1.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 2.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.6 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 21.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.8 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 3.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 3.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 3.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 2.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 2.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 91.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 1.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 1.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 2.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 2.0 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.9 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 15.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 6.8 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.1 | 3.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 2.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.7 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 5.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.9 | GO:0016248 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.1 | 1.6 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.8 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 4.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 1.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 2.0 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.1 | 0.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 1.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 4.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 3.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 1.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 5.1 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.1 | 0.2 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.1 | 0.2 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 3.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.3 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 1.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 3.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.5 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 1.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 2.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.5 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.2 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 4.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 2.1 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 5.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 2.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 3.0 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 1.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 1.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 5.1 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 4.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 5.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 83.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.8 | 54.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.6 | 19.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.6 | 18.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.4 | 13.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.4 | 18.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.4 | 26.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 12.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 6.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 6.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.2 | 58.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 12.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 4.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 1.9 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.2 | 8.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 9.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 32.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 6.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 3.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 9.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 5.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 6.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 1.6 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 1.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 25.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 2.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 2.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 2.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 15.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.7 | 18.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.6 | 72.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.5 | 12.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.4 | 8.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 27.4 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.4 | 12.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 5.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 20.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.3 | 5.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 0.5 | REACTOME P75NTR SIGNALS VIA NFKB | Genes involved in p75NTR signals via NF-kB |
| 0.3 | 13.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 9.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.2 | 3.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 4.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 6.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 5.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 1.2 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.2 | 3.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 6.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 5.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 2.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 5.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 4.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 2.5 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 5.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.9 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 6.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 4.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 3.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 9.0 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 1.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 2.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 3.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 3.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 4.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 17.1 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 6.0 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 2.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.9 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 3.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 0.8 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 1.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 2.2 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 4.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 5.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 2.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 1.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.1 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |