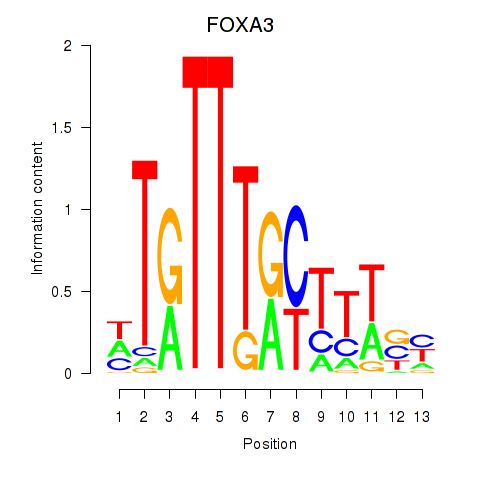
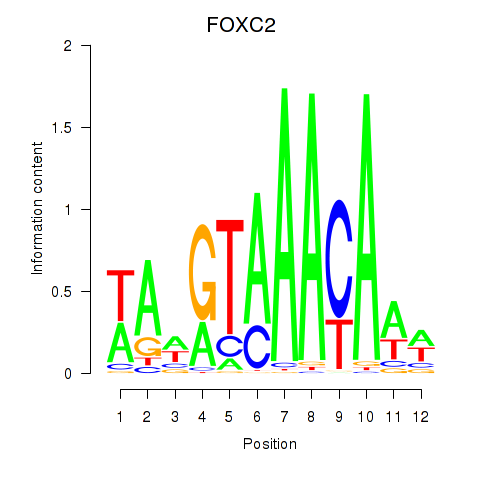
Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for FOXA3_FOXC2
Z-value: 0.51
Transcription factors associated with FOXA3_FOXC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA3
|
ENSG00000170608.2 | forkhead box A3 |
|
FOXC2
|
ENSG00000176692.4 | forkhead box C2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXC2 | hg19_v2_chr16_+_86600857_86600921 | 0.18 | 3.5e-01 | Click! |
| FOXA3 | hg19_v2_chr19_+_46367518_46367567 | -0.05 | 8.0e-01 | Click! |
Activity profile of FOXA3_FOXC2 motif
Sorted Z-values of FOXA3_FOXC2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_31823792 | 6.50 |
ENST00000375413.4
ENST00000354297.4 ENST00000375422.2 |
BPIFA1
|
BPI fold containing family A, member 1 |
| chr11_+_27076764 | 2.88 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr5_+_156712372 | 2.58 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr20_+_31870927 | 2.11 |
ENST00000253354.1
|
BPIFB1
|
BPI fold containing family B, member 1 |
| chr11_-_108464465 | 1.93 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr11_-_108464321 | 1.65 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chr7_+_142829162 | 1.52 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr13_+_50589390 | 1.17 |
ENST00000360473.4
ENST00000312942.1 |
KCNRG
|
potassium channel regulator |
| chr1_-_207095324 | 1.10 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr20_+_9049682 | 1.05 |
ENST00000334005.3
ENST00000378473.3 |
PLCB4
|
phospholipase C, beta 4 |
| chr1_-_146696901 | 1.05 |
ENST00000369272.3
ENST00000441068.2 |
FMO5
|
flavin containing monooxygenase 5 |
| chr17_+_79953310 | 1.02 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr13_-_86373536 | 0.98 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr17_+_72426891 | 0.97 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr1_+_104293028 | 0.84 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr13_-_36429763 | 0.81 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr4_+_165675197 | 0.81 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr9_-_75567962 | 0.78 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr10_-_21435488 | 0.74 |
ENST00000534331.1
ENST00000529198.1 ENST00000377118.4 |
C10orf113
|
chromosome 10 open reading frame 113 |
| chr13_-_39564993 | 0.74 |
ENST00000423210.1
|
STOML3
|
stomatin (EPB72)-like 3 |
| chrX_+_9431324 | 0.63 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr1_+_104159999 | 0.62 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr12_-_71551652 | 0.59 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr3_+_69985734 | 0.58 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr2_-_175711133 | 0.56 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr1_-_207095212 | 0.56 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr20_-_55841398 | 0.55 |
ENST00000395864.3
|
BMP7
|
bone morphogenetic protein 7 |
| chr4_+_165675269 | 0.54 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr10_+_95848824 | 0.54 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr3_-_45957088 | 0.52 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr1_+_61547405 | 0.52 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr20_+_52105495 | 0.51 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr12_+_59989918 | 0.48 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr3_-_107941209 | 0.46 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr18_+_3449330 | 0.46 |
ENST00000549253.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr15_+_71185148 | 0.44 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr12_-_76879852 | 0.43 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr14_+_56585048 | 0.41 |
ENST00000267460.4
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr3_-_45957534 | 0.41 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr1_-_201096312 | 0.41 |
ENST00000449188.2
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr11_-_116708302 | 0.40 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr1_+_87797351 | 0.39 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr11_+_6897856 | 0.38 |
ENST00000379829.2
|
OR10A4
|
olfactory receptor, family 10, subfamily A, member 4 |
| chr4_-_138453559 | 0.38 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr14_-_31926623 | 0.37 |
ENST00000356180.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr7_+_116654935 | 0.36 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr17_+_7487146 | 0.36 |
ENST00000396501.4
ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr20_-_55841662 | 0.35 |
ENST00000395863.3
ENST00000450594.2 |
BMP7
|
bone morphogenetic protein 7 |
| chr14_-_25479811 | 0.35 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr17_+_72427477 | 0.34 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr1_+_244214577 | 0.33 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr9_-_14722715 | 0.33 |
ENST00000380911.3
|
CER1
|
cerberus 1, DAN family BMP antagonist |
| chr6_-_41715128 | 0.32 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chr3_-_107941230 | 0.32 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr1_+_61548225 | 0.32 |
ENST00000371187.3
|
NFIA
|
nuclear factor I/A |
| chr1_-_223308098 | 0.32 |
ENST00000342210.6
|
TLR5
|
toll-like receptor 5 |
| chr19_+_42806250 | 0.30 |
ENST00000598490.1
ENST00000341747.3 |
PRR19
|
proline rich 19 |
| chr3_+_69985792 | 0.28 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr3_+_50712672 | 0.28 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr11_-_110561721 | 0.28 |
ENST00000357139.3
|
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr9_-_28670283 | 0.28 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr5_-_41213607 | 0.27 |
ENST00000337836.5
ENST00000433294.1 |
C6
|
complement component 6 |
| chrX_+_10124977 | 0.27 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr3_-_105588231 | 0.27 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr1_+_12806141 | 0.27 |
ENST00000288048.5
|
C1orf158
|
chromosome 1 open reading frame 158 |
| chr14_+_74034310 | 0.26 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr14_-_101036119 | 0.26 |
ENST00000355173.2
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr1_+_35225339 | 0.26 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr19_-_18902106 | 0.25 |
ENST00000542601.2
ENST00000425807.1 ENST00000222271.2 |
COMP
|
cartilage oligomeric matrix protein |
| chr1_+_18957500 | 0.25 |
ENST00000375375.3
|
PAX7
|
paired box 7 |
| chr5_+_40841276 | 0.25 |
ENST00000254691.5
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr16_-_4664860 | 0.25 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr12_-_39734783 | 0.25 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr16_-_4665023 | 0.24 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr12_-_92539614 | 0.24 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr9_-_3469181 | 0.24 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr22_+_46449674 | 0.23 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr18_-_53070913 | 0.23 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr10_+_124320156 | 0.22 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr2_-_188312971 | 0.22 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr9_+_70856397 | 0.22 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr2_+_179318295 | 0.22 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr2_+_175260451 | 0.22 |
ENST00000458563.1
ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3
|
secernin 3 |
| chr1_+_114471809 | 0.21 |
ENST00000426820.2
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr10_+_124320195 | 0.21 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr4_+_187148556 | 0.21 |
ENST00000264690.6
ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1 |
| chr14_-_70263979 | 0.21 |
ENST00000216540.4
|
SLC10A1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr8_+_19171128 | 0.20 |
ENST00000265807.3
|
SH2D4A
|
SH2 domain containing 4A |
| chr4_-_70080449 | 0.20 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr9_-_70490107 | 0.20 |
ENST00000377395.4
ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5
|
COBW domain containing 5 |
| chr7_+_134832808 | 0.20 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr11_-_107582775 | 0.20 |
ENST00000305991.2
|
SLN
|
sarcolipin |
| chr3_-_193096600 | 0.19 |
ENST00000446087.1
ENST00000342358.4 |
ATP13A5
|
ATPase type 13A5 |
| chr4_-_11431389 | 0.19 |
ENST00000002596.5
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr17_+_4613776 | 0.19 |
ENST00000269260.2
|
ARRB2
|
arrestin, beta 2 |
| chr15_+_60296421 | 0.19 |
ENST00000396057.4
|
FOXB1
|
forkhead box B1 |
| chr10_-_49482907 | 0.19 |
ENST00000374201.3
ENST00000407470.4 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr14_+_105267250 | 0.19 |
ENST00000342537.7
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr15_+_96869165 | 0.19 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_+_135596180 | 0.19 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr13_+_52598827 | 0.19 |
ENST00000521776.2
|
UTP14C
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) |
| chr1_+_29563011 | 0.19 |
ENST00000345512.3
ENST00000373779.3 ENST00000356870.3 ENST00000323874.8 ENST00000428026.2 ENST00000460170.2 |
PTPRU
|
protein tyrosine phosphatase, receptor type, U |
| chr16_-_15474904 | 0.18 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr1_+_101361782 | 0.18 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr17_+_4613918 | 0.18 |
ENST00000574954.1
ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2
|
arrestin, beta 2 |
| chr12_+_59989791 | 0.18 |
ENST00000552432.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr16_+_69958887 | 0.18 |
ENST00000568684.1
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr4_+_86396265 | 0.18 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr3_+_119316689 | 0.18 |
ENST00000273371.4
|
PLA1A
|
phospholipase A1 member A |
| chr12_-_92536433 | 0.17 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr2_+_135596106 | 0.17 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr3_+_119316721 | 0.17 |
ENST00000488919.1
ENST00000495992.1 |
PLA1A
|
phospholipase A1 member A |
| chr17_-_26733179 | 0.17 |
ENST00000440501.1
ENST00000321666.5 |
SLC46A1
|
solute carrier family 46 (folate transporter), member 1 |
| chr16_-_3350614 | 0.17 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr14_-_31926701 | 0.16 |
ENST00000310850.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chrX_+_24167746 | 0.16 |
ENST00000428571.1
ENST00000539115.1 |
ZFX
|
zinc finger protein, X-linked |
| chr4_+_86396321 | 0.16 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr17_+_8924837 | 0.16 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr15_+_28624878 | 0.16 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr5_+_140787600 | 0.15 |
ENST00000520790.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6 |
| chr1_-_246729544 | 0.15 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr19_+_47421933 | 0.15 |
ENST00000404338.3
|
ARHGAP35
|
Rho GTPase activating protein 35 |
| chr4_-_140223670 | 0.15 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr4_-_140223614 | 0.15 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr3_-_187455680 | 0.15 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr21_-_43786634 | 0.15 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chr14_-_24664540 | 0.15 |
ENST00000530563.1
ENST00000528895.1 ENST00000528669.1 ENST00000532632.1 |
TM9SF1
|
transmembrane 9 superfamily member 1 |
| chr10_+_123923105 | 0.15 |
ENST00000368999.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr16_+_14802801 | 0.15 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr8_+_95908041 | 0.14 |
ENST00000396113.1
ENST00000519136.1 |
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr22_-_30867973 | 0.14 |
ENST00000402286.1
ENST00000401751.1 ENST00000539629.1 ENST00000403066.1 ENST00000215812.4 |
SEC14L3
|
SEC14-like 3 (S. cerevisiae) |
| chrX_+_65382381 | 0.14 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr1_+_246729724 | 0.14 |
ENST00000366513.4
ENST00000366512.3 |
CNST
|
consortin, connexin sorting protein |
| chr3_-_114343039 | 0.14 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr3_+_148545586 | 0.14 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chrX_+_135730373 | 0.14 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr1_+_74701062 | 0.14 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr13_+_73629107 | 0.13 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chrX_+_24167828 | 0.13 |
ENST00000379188.3
ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX
|
zinc finger protein, X-linked |
| chr6_+_21666633 | 0.13 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr4_+_146403912 | 0.13 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr1_+_239882842 | 0.13 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr14_+_56127989 | 0.13 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr6_+_52051171 | 0.13 |
ENST00000340057.1
|
IL17A
|
interleukin 17A |
| chr1_-_150602035 | 0.13 |
ENST00000503241.1
ENST00000369016.4 ENST00000339643.5 ENST00000271690.8 ENST00000356527.5 ENST00000362052.7 ENST00000503345.1 ENST00000369014.5 ENST00000369009.3 |
ENSA
|
endosulfine alpha |
| chr19_-_46088068 | 0.13 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr8_+_95907993 | 0.13 |
ENST00000523378.1
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr2_-_32490859 | 0.13 |
ENST00000404025.2
|
NLRC4
|
NLR family, CARD domain containing 4 |
| chr15_-_40600026 | 0.13 |
ENST00000456256.2
ENST00000557821.1 |
PLCB2
|
phospholipase C, beta 2 |
| chr2_-_227050079 | 0.13 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chrX_+_135730297 | 0.13 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr5_+_161494770 | 0.13 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr1_-_43855479 | 0.12 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr20_+_43935474 | 0.12 |
ENST00000372743.1
ENST00000372741.3 ENST00000343694.3 |
RBPJL
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr11_-_102401469 | 0.12 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr3_+_132316081 | 0.12 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr5_+_161494521 | 0.12 |
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr3_+_171561127 | 0.12 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr6_-_159466042 | 0.12 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr13_-_99910673 | 0.12 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chr13_+_95364963 | 0.12 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chr6_+_161123270 | 0.12 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr19_-_47157914 | 0.12 |
ENST00000300875.4
|
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr12_-_122018346 | 0.12 |
ENST00000377069.4
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr8_-_133772794 | 0.12 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr17_-_39183452 | 0.12 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr22_+_35462129 | 0.12 |
ENST00000404699.2
ENST00000308700.6 |
ISX
|
intestine-specific homeobox |
| chr3_+_114012819 | 0.12 |
ENST00000383671.3
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr3_+_108855558 | 0.11 |
ENST00000467240.1
ENST00000477643.1 ENST00000479039.1 ENST00000593799.1 |
RP11-59E19.1
|
RP11-59E19.1 |
| chr1_+_43855560 | 0.11 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr6_+_44310421 | 0.11 |
ENST00000288390.2
|
SPATS1
|
spermatogenesis associated, serine-rich 1 |
| chr17_-_6947225 | 0.11 |
ENST00000574600.1
ENST00000308009.1 ENST00000447225.1 |
SLC16A11
|
solute carrier family 16, member 11 |
| chr12_-_15038779 | 0.11 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr14_-_21562648 | 0.11 |
ENST00000555270.1
|
ZNF219
|
zinc finger protein 219 |
| chr6_-_159466136 | 0.11 |
ENST00000367066.3
ENST00000326965.6 |
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr1_+_159141397 | 0.11 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr1_-_150669500 | 0.11 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr14_-_74551096 | 0.10 |
ENST00000350259.4
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr14_-_36988882 | 0.10 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr16_+_20775358 | 0.10 |
ENST00000440284.2
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr1_+_154540246 | 0.10 |
ENST00000368476.3
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal) |
| chrX_-_133792480 | 0.10 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr10_-_31288398 | 0.10 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chrX_-_80457385 | 0.10 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr12_+_58087901 | 0.10 |
ENST00000315970.7
ENST00000547079.1 ENST00000439210.2 ENST00000389146.6 ENST00000413095.2 ENST00000551035.1 ENST00000257966.8 ENST00000435406.2 ENST00000550372.1 ENST00000389142.5 |
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr12_-_48099754 | 0.10 |
ENST00000380650.4
|
RPAP3
|
RNA polymerase II associated protein 3 |
| chr1_+_114471972 | 0.10 |
ENST00000369559.4
ENST00000369554.2 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr6_+_44310376 | 0.10 |
ENST00000515220.1
ENST00000323108.8 |
SPATS1
|
spermatogenesis associated, serine-rich 1 |
| chr4_-_88450244 | 0.09 |
ENST00000503414.1
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr12_+_48516357 | 0.09 |
ENST00000549022.1
ENST00000547587.1 ENST00000312352.7 |
PFKM
|
phosphofructokinase, muscle |
| chr1_-_150669604 | 0.09 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr8_+_52730143 | 0.09 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr3_+_137728842 | 0.09 |
ENST00000183605.5
|
CLDN18
|
claudin 18 |
| chr17_-_79212825 | 0.09 |
ENST00000374769.2
|
ENTHD2
|
ENTH domain containing 2 |
| chr17_-_26697304 | 0.09 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr12_-_48099773 | 0.09 |
ENST00000432584.3
ENST00000005386.3 |
RPAP3
|
RNA polymerase II associated protein 3 |
| chr2_-_225434538 | 0.09 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr1_+_150488205 | 0.08 |
ENST00000416894.1
|
LINC00568
|
long intergenic non-protein coding RNA 568 |
| chr4_+_187187098 | 0.08 |
ENST00000403665.2
ENST00000264692.4 |
F11
|
coagulation factor XI |
| chr12_-_18243119 | 0.08 |
ENST00000538724.1
ENST00000229002.2 |
RERGL
|
RERG/RAS-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA3_FOXC2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.5 | GO:0044010 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.3 | 0.9 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.3 | 2.1 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.3 | 2.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 2.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 0.9 | GO:1900106 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme morphogenesis(GO:0072134) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) allantois development(GO:1905069) |
| 0.2 | 0.8 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 1.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 0.3 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.2 | 3.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.3 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.4 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.1 | 0.6 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.7 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.4 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 0.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 1.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.3 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.1 | 0.1 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 | 0.2 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.3 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.3 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.4 | GO:0034627 | quinolinate biosynthetic process(GO:0019805) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.1 | 1.5 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.1 | 0.2 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.0 | 0.7 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.2 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.4 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.0 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.8 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.4 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) |
| 0.0 | 0.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.9 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 1.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.2 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.3 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.9 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.0 | GO:0060405 | regulation of penile erection(GO:0060405) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.3 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 1.3 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.3 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.0 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.1 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.3 | GO:0003417 | growth plate cartilage development(GO:0003417) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.2 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 0.8 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.2 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 0.5 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.2 | 0.7 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 0.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.2 | 0.6 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 1.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.4 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.1 | 0.4 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 1.5 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.3 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.3 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 1.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.3 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.4 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 3.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 2.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |