Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
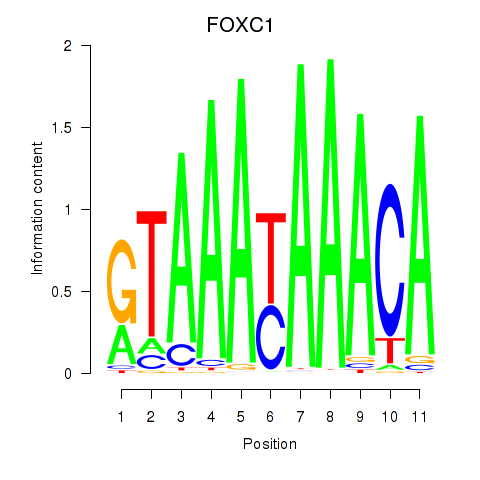
Results for FOXC1
Z-value: 0.62
Transcription factors associated with FOXC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXC1
|
ENSG00000054598.5 | forkhead box C1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXC1 | hg19_v2_chr6_+_1610681_1610681 | 0.39 | 3.4e-02 | Click! |
Activity profile of FOXC1 motif
Sorted Z-values of FOXC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_72375167 | 3.48 |
ENST00000001146.2
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr8_-_23261589 | 2.14 |
ENST00000524168.1
ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2
|
lysyl oxidase-like 2 |
| chr2_-_161056762 | 1.46 |
ENST00000428609.2
ENST00000409967.2 |
ITGB6
|
integrin, beta 6 |
| chr2_-_161056802 | 1.28 |
ENST00000283249.2
ENST00000409872.1 |
ITGB6
|
integrin, beta 6 |
| chr13_-_61989655 | 1.25 |
ENST00000409204.4
|
PCDH20
|
protocadherin 20 |
| chr8_+_54764346 | 1.10 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr21_+_39628852 | 0.92 |
ENST00000398938.2
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr3_-_16524357 | 0.88 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr14_+_37126765 | 0.84 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr4_-_103266626 | 0.82 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr1_+_12524965 | 0.75 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr10_+_105314881 | 0.71 |
ENST00000437579.1
|
NEURL
|
neuralized E3 ubiquitin protein ligase 1 |
| chr17_-_37764128 | 0.69 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr22_+_46449674 | 0.69 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr14_-_57272366 | 0.67 |
ENST00000554788.1
ENST00000554845.1 ENST00000408990.3 |
OTX2
|
orthodenticle homeobox 2 |
| chr9_-_89562104 | 0.63 |
ENST00000298743.7
|
GAS1
|
growth arrest-specific 1 |
| chr14_+_76776957 | 0.63 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr1_-_94079648 | 0.62 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr6_+_34204642 | 0.61 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr17_-_57229155 | 0.59 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr10_+_71561704 | 0.58 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr10_+_54074033 | 0.58 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr7_-_99698338 | 0.56 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr8_+_11666649 | 0.54 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr1_-_109935819 | 0.54 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr19_-_51141196 | 0.50 |
ENST00000338916.4
|
SYT3
|
synaptotagmin III |
| chr2_+_113885138 | 0.50 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr18_+_60382672 | 0.46 |
ENST00000400316.4
ENST00000262719.5 |
PHLPP1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr8_+_70404996 | 0.45 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr15_+_76016293 | 0.44 |
ENST00000332145.2
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1 |
| chr4_-_99578789 | 0.43 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr8_-_134309335 | 0.43 |
ENST00000522890.1
ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1
|
N-myc downstream regulated 1 |
| chr12_-_99038732 | 0.42 |
ENST00000393042.3
ENST00000420861.1 ENST00000299157.4 ENST00000342502.2 |
IKBIP
|
IKBKB interacting protein |
| chr8_-_134309823 | 0.41 |
ENST00000414097.2
|
NDRG1
|
N-myc downstream regulated 1 |
| chr20_-_22566089 | 0.41 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr7_+_129906660 | 0.41 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr4_-_99578776 | 0.39 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr1_-_94312706 | 0.37 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr21_+_39628655 | 0.37 |
ENST00000398925.1
ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr2_-_190044480 | 0.36 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr5_-_127674883 | 0.36 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr10_+_71561649 | 0.36 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr3_-_71632894 | 0.35 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr5_-_115872142 | 0.34 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr12_+_43086018 | 0.34 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr11_-_121986923 | 0.34 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr13_+_98086445 | 0.33 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr4_-_89744457 | 0.33 |
ENST00000395002.2
|
FAM13A
|
family with sequence similarity 13, member A |
| chr19_+_36024310 | 0.33 |
ENST00000222286.4
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr10_+_105315102 | 0.32 |
ENST00000369777.2
|
NEURL
|
neuralized E3 ubiquitin protein ligase 1 |
| chr1_+_150122034 | 0.32 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chr18_+_19749386 | 0.31 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr1_-_28384598 | 0.30 |
ENST00000373864.1
|
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chr4_-_164534657 | 0.30 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr16_+_8715574 | 0.30 |
ENST00000561758.1
|
METTL22
|
methyltransferase like 22 |
| chr5_+_161494521 | 0.30 |
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr8_-_108510224 | 0.30 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr16_+_8715536 | 0.30 |
ENST00000563958.1
ENST00000381920.3 ENST00000564554.1 |
METTL22
|
methyltransferase like 22 |
| chr8_+_31497271 | 0.29 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr18_+_29171689 | 0.28 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr16_+_30064411 | 0.28 |
ENST00000338110.5
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr10_+_114710425 | 0.28 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr21_+_38071430 | 0.28 |
ENST00000290399.6
|
SIM2
|
single-minded family bHLH transcription factor 2 |
| chr16_+_30064444 | 0.27 |
ENST00000395248.1
ENST00000566897.1 ENST00000568435.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr6_+_47666275 | 0.27 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr8_-_72274467 | 0.27 |
ENST00000340726.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr5_+_161494770 | 0.27 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr11_+_112832090 | 0.26 |
ENST00000533760.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chrX_+_85969626 | 0.26 |
ENST00000484479.1
|
DACH2
|
dachshund homolog 2 (Drosophila) |
| chr6_+_32121908 | 0.26 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr3_-_178865747 | 0.26 |
ENST00000435560.1
|
RP11-360P21.2
|
RP11-360P21.2 |
| chr8_-_72274095 | 0.26 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr20_-_62587735 | 0.25 |
ENST00000354216.6
ENST00000369892.3 ENST00000358711.3 |
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr12_-_53994805 | 0.25 |
ENST00000328463.7
|
ATF7
|
activating transcription factor 7 |
| chr2_+_105050794 | 0.25 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr2_+_33359687 | 0.25 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr12_+_93096759 | 0.25 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr2_-_208030647 | 0.25 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr7_+_139529040 | 0.25 |
ENST00000455353.1
ENST00000458722.1 ENST00000411653.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr15_+_63188009 | 0.25 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chrX_+_99839799 | 0.25 |
ENST00000373031.4
|
TNMD
|
tenomodulin |
| chr12_+_99038998 | 0.24 |
ENST00000359972.2
ENST00000357310.1 ENST00000339433.3 ENST00000333991.1 |
APAF1
|
apoptotic peptidase activating factor 1 |
| chr2_+_33359646 | 0.24 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr12_+_93096619 | 0.24 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr1_-_43855444 | 0.24 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr6_+_32121789 | 0.24 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr1_+_223101757 | 0.24 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr4_-_74486347 | 0.23 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr14_-_24020858 | 0.23 |
ENST00000419474.3
|
ZFHX2
|
zinc finger homeobox 2 |
| chr14_+_100070869 | 0.23 |
ENST00000502101.2
|
RP11-543C4.1
|
RP11-543C4.1 |
| chr10_-_4285923 | 0.23 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr8_+_97597148 | 0.23 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr12_+_99038919 | 0.23 |
ENST00000551964.1
|
APAF1
|
apoptotic peptidase activating factor 1 |
| chr15_+_67418047 | 0.23 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr8_-_93107696 | 0.23 |
ENST00000436581.2
ENST00000520583.1 ENST00000519061.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr3_+_171758344 | 0.22 |
ENST00000336824.4
ENST00000423424.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr15_-_72563585 | 0.22 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr7_+_136553370 | 0.22 |
ENST00000445907.2
|
CHRM2
|
cholinergic receptor, muscarinic 2 |
| chr2_+_210517895 | 0.22 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr2_+_28974668 | 0.21 |
ENST00000296122.6
ENST00000395366.2 |
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr4_-_74486109 | 0.21 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr13_-_36050819 | 0.21 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr4_+_15376165 | 0.21 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr7_+_139528952 | 0.21 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr12_+_59989918 | 0.21 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr8_-_95449155 | 0.21 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr20_-_35890211 | 0.20 |
ENST00000373614.2
|
GHRH
|
growth hormone releasing hormone |
| chr4_+_71587669 | 0.20 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr10_+_50507181 | 0.20 |
ENST00000323868.4
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr8_-_19540086 | 0.20 |
ENST00000332246.6
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr6_-_42418999 | 0.20 |
ENST00000340840.2
ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chr2_+_28974603 | 0.20 |
ENST00000441461.1
ENST00000358506.2 |
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr16_-_740354 | 0.20 |
ENST00000293883.4
|
WDR24
|
WD repeat domain 24 |
| chr16_-_740419 | 0.19 |
ENST00000248142.6
|
WDR24
|
WD repeat domain 24 |
| chr9_+_6413317 | 0.19 |
ENST00000276893.5
ENST00000381373.3 |
UHRF2
|
ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase |
| chr3_-_168865522 | 0.19 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr22_-_38699003 | 0.19 |
ENST00000451964.1
|
CSNK1E
|
casein kinase 1, epsilon |
| chr10_+_50507232 | 0.19 |
ENST00000374144.3
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr13_-_99667960 | 0.19 |
ENST00000448493.2
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr5_+_159614374 | 0.18 |
ENST00000393980.4
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr1_+_199996702 | 0.18 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr10_+_71561630 | 0.18 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr6_-_100442077 | 0.18 |
ENST00000281806.2
ENST00000369212.2 |
MCHR2
|
melanin-concentrating hormone receptor 2 |
| chr5_-_36241900 | 0.18 |
ENST00000381937.4
ENST00000514504.1 |
NADK2
|
NAD kinase 2, mitochondrial |
| chr10_+_114709999 | 0.18 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr7_-_72972319 | 0.18 |
ENST00000223368.2
|
BCL7B
|
B-cell CLL/lymphoma 7B |
| chr19_+_17516909 | 0.17 |
ENST00000601007.1
ENST00000594913.1 ENST00000599975.1 ENST00000600514.1 |
CTD-2521M24.9
MVB12A
|
CTD-2521M24.9 multivesicular body subunit 12A |
| chr6_+_119215308 | 0.17 |
ENST00000229595.5
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr10_-_25010795 | 0.17 |
ENST00000416305.1
ENST00000376410.2 |
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr12_+_16109519 | 0.17 |
ENST00000526530.1
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr8_-_80993010 | 0.17 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr2_-_32490859 | 0.17 |
ENST00000404025.2
|
NLRC4
|
NLR family, CARD domain containing 4 |
| chr4_-_186696425 | 0.16 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr19_+_29493456 | 0.16 |
ENST00000591143.1
ENST00000592653.1 |
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr8_-_145754428 | 0.16 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr20_-_62582475 | 0.16 |
ENST00000369908.5
|
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr6_-_32122106 | 0.15 |
ENST00000428778.1
|
PRRT1
|
proline-rich transmembrane protein 1 |
| chr6_+_84563295 | 0.15 |
ENST00000369687.1
|
RIPPLY2
|
ripply transcriptional repressor 2 |
| chr3_+_57875738 | 0.15 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr12_-_76462713 | 0.15 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr17_-_27503770 | 0.14 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr11_-_66112555 | 0.14 |
ENST00000425825.2
ENST00000359957.3 |
BRMS1
|
breast cancer metastasis suppressor 1 |
| chr3_+_189349162 | 0.14 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr2_-_72374948 | 0.14 |
ENST00000546307.1
ENST00000474509.1 |
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr3_-_133969673 | 0.14 |
ENST00000427044.2
|
RYK
|
receptor-like tyrosine kinase |
| chr17_+_41363854 | 0.14 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr19_-_40919271 | 0.14 |
ENST00000291825.7
ENST00000324001.7 |
PRX
|
periaxin |
| chr12_-_31479107 | 0.14 |
ENST00000542983.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr8_-_93107827 | 0.14 |
ENST00000520724.1
ENST00000518844.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr3_-_113897545 | 0.14 |
ENST00000467632.1
|
DRD3
|
dopamine receptor D3 |
| chr8_-_141931287 | 0.14 |
ENST00000517887.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr3_+_178866199 | 0.14 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr5_+_161495038 | 0.14 |
ENST00000393933.4
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr12_-_92539614 | 0.13 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr7_+_80231466 | 0.13 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr1_-_159684371 | 0.13 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr1_+_18958008 | 0.13 |
ENST00000420770.2
ENST00000400661.3 |
PAX7
|
paired box 7 |
| chr1_-_153931052 | 0.13 |
ENST00000368630.3
ENST00000368633.1 |
CRTC2
|
CREB regulated transcription coactivator 2 |
| chr1_+_197886461 | 0.13 |
ENST00000367388.3
ENST00000337020.2 ENST00000367387.4 |
LHX9
|
LIM homeobox 9 |
| chr5_-_36242119 | 0.13 |
ENST00000511088.1
ENST00000282512.3 ENST00000506945.1 |
NADK2
|
NAD kinase 2, mitochondrial |
| chr10_-_97200772 | 0.12 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr4_+_147096837 | 0.12 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr14_+_69726656 | 0.12 |
ENST00000337827.4
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr21_+_17792672 | 0.12 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr8_+_31496809 | 0.12 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr15_-_30114622 | 0.12 |
ENST00000495972.2
ENST00000346128.6 |
TJP1
|
tight junction protein 1 |
| chrX_-_77914825 | 0.12 |
ENST00000321110.1
|
ZCCHC5
|
zinc finger, CCHC domain containing 5 |
| chr1_-_149982624 | 0.12 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr18_-_52626622 | 0.11 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr6_+_10528560 | 0.11 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr10_+_114710211 | 0.11 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr4_+_86396321 | 0.11 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr1_+_234509186 | 0.11 |
ENST00000366615.4
|
COA6
|
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr4_-_83765613 | 0.11 |
ENST00000503937.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr4_+_71588372 | 0.11 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr19_-_36523529 | 0.11 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr10_+_71562180 | 0.11 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr6_+_108882069 | 0.10 |
ENST00000406360.1
|
FOXO3
|
forkhead box O3 |
| chr12_-_123756781 | 0.10 |
ENST00000544658.1
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr3_+_46449049 | 0.10 |
ENST00000357392.4
ENST00000400880.3 ENST00000433848.1 |
CCRL2
|
chemokine (C-C motif) receptor-like 2 |
| chr3_-_48481518 | 0.10 |
ENST00000412398.2
ENST00000395696.1 |
CCDC51
|
coiled-coil domain containing 51 |
| chr6_-_42016385 | 0.10 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr19_-_56343353 | 0.10 |
ENST00000592953.1
ENST00000589093.1 |
NLRP11
|
NLR family, pyrin domain containing 11 |
| chr6_+_134274322 | 0.10 |
ENST00000367871.1
ENST00000237264.4 |
TBPL1
|
TBP-like 1 |
| chr3_-_49066811 | 0.10 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr12_-_31479045 | 0.10 |
ENST00000539409.1
ENST00000395766.1 |
FAM60A
|
family with sequence similarity 60, member A |
| chr12_+_54348618 | 0.09 |
ENST00000243103.3
|
HOXC12
|
homeobox C12 |
| chr3_+_57875711 | 0.09 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr15_-_65321943 | 0.09 |
ENST00000220058.4
|
MTFMT
|
mitochondrial methionyl-tRNA formyltransferase |
| chr3_-_48481434 | 0.09 |
ENST00000395694.2
ENST00000447018.1 ENST00000442740.1 |
CCDC51
|
coiled-coil domain containing 51 |
| chr6_-_32083106 | 0.09 |
ENST00000442721.1
|
TNXB
|
tenascin XB |
| chr12_+_59989791 | 0.09 |
ENST00000552432.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_-_169703203 | 0.09 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr3_-_49131788 | 0.09 |
ENST00000395443.2
ENST00000411682.1 |
QRICH1
|
glutamine-rich 1 |
| chr3_-_141868293 | 0.09 |
ENST00000317104.7
ENST00000494358.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr18_+_32073253 | 0.09 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr5_+_135468516 | 0.08 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chrX_-_117107680 | 0.08 |
ENST00000447671.2
ENST00000262820.3 |
KLHL13
|
kelch-like family member 13 |
| chr20_-_17511962 | 0.08 |
ENST00000377873.3
|
BFSP1
|
beaded filament structural protein 1, filensin |
| chr19_-_4831701 | 0.08 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:1902811 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.2 | 2.7 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 0.7 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.2 | 0.6 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.2 | 0.6 | GO:1904956 | regulation of endodermal cell fate specification(GO:0042663) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.2 | 0.4 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.4 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.6 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.7 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.1 | 0.9 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.5 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.6 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.3 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.9 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.8 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.3 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.5 | GO:0072423 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.1 | 0.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.2 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.3 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.1 | 0.5 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 2.1 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 0.5 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.4 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.5 | GO:0002249 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.1 | 0.5 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.6 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.1 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 1.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 1.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.0 | 0.8 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.1 | GO:1904760 | myofibroblast differentiation(GO:0036446) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.4 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0051194 | positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.0 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.3 | 1.2 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.5 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.3 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.6 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 1.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.4 | 2.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 0.5 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.2 | 0.5 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.5 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.5 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 1.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.4 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 0.2 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 0.2 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 0.2 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.1 | 0.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.6 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 3.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.8 | GO:0016279 | protein-lysine N-methyltransferase activity(GO:0016279) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 4.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |