Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
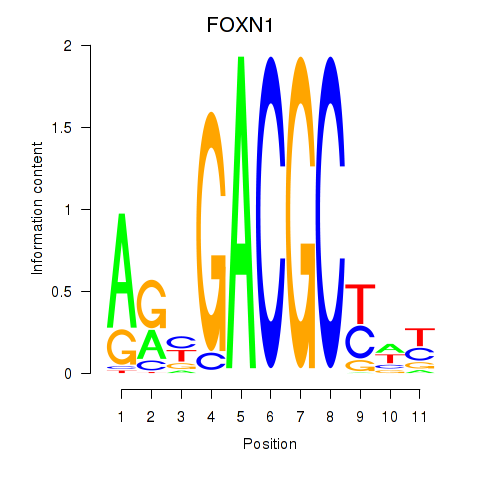
Results for FOXN1
Z-value: 0.78
Transcription factors associated with FOXN1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXN1
|
ENSG00000109101.3 | forkhead box N1 |
Activity profile of FOXN1 motif
Sorted Z-values of FOXN1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_28113217 | 4.95 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr15_+_71184931 | 3.19 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr4_+_81256871 | 3.13 |
ENST00000358105.3
ENST00000508675.1 |
C4orf22
|
chromosome 4 open reading frame 22 |
| chr1_+_245133656 | 3.11 |
ENST00000366521.3
|
EFCAB2
|
EF-hand calcium binding domain 2 |
| chr15_+_71185148 | 2.82 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr9_+_34458771 | 2.53 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr5_+_76506706 | 2.44 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr7_+_76751926 | 2.43 |
ENST00000285871.4
ENST00000431197.1 |
CCDC146
|
coiled-coil domain containing 146 |
| chr17_+_62075703 | 2.33 |
ENST00000577953.1
ENST00000582540.1 ENST00000579184.1 ENST00000425164.3 ENST00000412177.1 ENST00000539996.1 ENST00000583891.1 ENST00000580752.1 |
C17orf72
|
chromosome 17 open reading frame 72 |
| chr14_+_65016620 | 2.19 |
ENST00000298705.1
|
PPP1R36
|
protein phosphatase 1, regulatory subunit 36 |
| chr5_-_100238956 | 2.07 |
ENST00000231461.5
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chrY_+_22737678 | 2.00 |
ENST00000382772.3
|
EIF1AY
|
eukaryotic translation initiation factor 1A, Y-linked |
| chr17_+_11501748 | 1.92 |
ENST00000262442.4
ENST00000579828.1 |
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr19_+_14544099 | 1.85 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr7_-_120498357 | 1.78 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr9_-_99381660 | 1.75 |
ENST00000375240.3
ENST00000463569.1 |
CDC14B
|
cell division cycle 14B |
| chr15_+_78556809 | 1.63 |
ENST00000343789.3
ENST00000394852.3 |
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr10_+_104180580 | 1.60 |
ENST00000425536.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr22_-_31741757 | 1.56 |
ENST00000215919.3
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr2_-_28113965 | 1.45 |
ENST00000302188.3
|
RBKS
|
ribokinase |
| chr9_-_99382065 | 1.44 |
ENST00000265659.2
ENST00000375241.1 ENST00000375236.1 |
CDC14B
|
cell division cycle 14B |
| chr2_+_73612858 | 1.33 |
ENST00000409009.1
ENST00000264448.6 ENST00000377715.1 |
ALMS1
|
Alstrom syndrome 1 |
| chr10_+_127585093 | 1.33 |
ENST00000368695.1
ENST00000368693.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr3_-_129147432 | 1.32 |
ENST00000503957.1
ENST00000505956.1 ENST00000326085.3 |
EFCAB12
|
EF-hand calcium binding domain 12 |
| chr3_-_27410847 | 1.22 |
ENST00000429845.2
ENST00000341435.5 ENST00000435750.1 |
NEK10
|
NIMA-related kinase 10 |
| chr20_-_3154162 | 1.14 |
ENST00000360342.3
|
LZTS3
|
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr5_+_140019004 | 1.11 |
ENST00000394671.3
ENST00000511410.1 ENST00000537378.1 |
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chrX_-_117107680 | 1.10 |
ENST00000447671.2
ENST00000262820.3 |
KLHL13
|
kelch-like family member 13 |
| chr1_-_59012365 | 1.09 |
ENST00000456980.1
ENST00000482274.2 ENST00000453710.1 ENST00000419242.1 ENST00000358603.2 ENST00000371226.3 ENST00000426139.1 |
OMA1
|
OMA1 zinc metallopeptidase |
| chr17_-_56065484 | 1.08 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr14_+_76452090 | 1.06 |
ENST00000314067.6
ENST00000238628.6 ENST00000556742.1 |
IFT43
|
intraflagellar transport 43 homolog (Chlamydomonas) |
| chr14_-_92302784 | 1.05 |
ENST00000340892.5
ENST00000360594.5 |
TC2N
|
tandem C2 domains, nuclear |
| chr1_-_114302086 | 1.05 |
ENST00000369604.1
ENST00000357783.2 |
PHTF1
|
putative homeodomain transcription factor 1 |
| chr3_+_93698974 | 1.05 |
ENST00000535334.1
ENST00000478400.1 ENST00000303097.7 ENST00000394222.3 ENST00000471138.1 ENST00000539730.1 |
ARL13B
|
ADP-ribosylation factor-like 13B |
| chr11_-_108464321 | 1.04 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chrX_-_117107542 | 1.04 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr1_-_114301960 | 0.99 |
ENST00000369598.1
ENST00000369600.1 |
PHTF1
|
putative homeodomain transcription factor 1 |
| chrX_-_108868390 | 0.97 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr1_-_114301503 | 0.97 |
ENST00000447664.2
|
PHTF1
|
putative homeodomain transcription factor 1 |
| chr5_+_140019079 | 0.96 |
ENST00000252100.6
|
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chr1_-_114301755 | 0.93 |
ENST00000393357.2
ENST00000369596.2 ENST00000446739.1 |
PHTF1
|
putative homeodomain transcription factor 1 |
| chr19_-_46389359 | 0.92 |
ENST00000302165.3
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1 |
| chr12_+_56473939 | 0.92 |
ENST00000450146.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr12_+_50451331 | 0.89 |
ENST00000228468.4
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr13_+_73356197 | 0.88 |
ENST00000326291.6
|
PIBF1
|
progesterone immunomodulatory binding factor 1 |
| chr3_-_58572760 | 0.87 |
ENST00000447756.2
|
FAM107A
|
family with sequence similarity 107, member A |
| chr6_+_29910301 | 0.87 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr14_-_92302825 | 0.86 |
ENST00000556018.1
|
TC2N
|
tandem C2 domains, nuclear |
| chr1_-_217311090 | 0.83 |
ENST00000493603.1
ENST00000366940.2 |
ESRRG
|
estrogen-related receptor gamma |
| chr16_-_30773372 | 0.83 |
ENST00000545825.1
ENST00000541260.1 |
C16orf93
|
chromosome 16 open reading frame 93 |
| chr6_+_33422343 | 0.83 |
ENST00000395064.2
|
ZBTB9
|
zinc finger and BTB domain containing 9 |
| chr13_-_24463530 | 0.76 |
ENST00000382172.3
|
MIPEP
|
mitochondrial intermediate peptidase |
| chr21_+_33784670 | 0.75 |
ENST00000300255.2
|
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr12_+_56473910 | 0.73 |
ENST00000411731.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr15_+_55700741 | 0.73 |
ENST00000569691.1
|
C15orf65
|
chromosome 15 open reading frame 65 |
| chr5_-_77656175 | 0.69 |
ENST00000513755.1
ENST00000421004.3 |
CTD-2037K23.2
|
CTD-2037K23.2 |
| chr1_+_101361782 | 0.69 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr10_-_104179682 | 0.67 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr5_+_149340282 | 0.66 |
ENST00000286298.4
|
SLC26A2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr12_-_102224704 | 0.66 |
ENST00000299314.7
|
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr10_-_52383644 | 0.66 |
ENST00000361781.2
|
SGMS1
|
sphingomyelin synthase 1 |
| chr15_+_92937144 | 0.65 |
ENST00000539113.1
ENST00000555434.1 |
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr1_+_171283331 | 0.62 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr13_-_88323218 | 0.61 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr20_+_3801162 | 0.61 |
ENST00000379573.2
ENST00000379567.2 ENST00000455742.1 ENST00000246041.2 |
AP5S1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr4_-_170533723 | 0.60 |
ENST00000510533.1
ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1
|
NIMA-related kinase 1 |
| chr17_+_7155556 | 0.59 |
ENST00000570500.1
ENST00000574993.1 ENST00000396628.2 ENST00000573657.1 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr15_+_34261089 | 0.59 |
ENST00000383263.5
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr3_-_52443799 | 0.58 |
ENST00000470173.1
ENST00000296288.5 |
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr2_-_110962544 | 0.57 |
ENST00000355301.4
ENST00000445609.2 ENST00000417665.1 ENST00000418527.1 ENST00000316534.4 ENST00000393272.3 |
NPHP1
|
nephronophthisis 1 (juvenile) |
| chr9_-_15307200 | 0.53 |
ENST00000506891.1
ENST00000541445.1 ENST00000512701.2 ENST00000380850.4 ENST00000297615.5 ENST00000355694.2 |
TTC39B
|
tetratricopeptide repeat domain 39B |
| chr2_-_242576864 | 0.53 |
ENST00000407315.1
|
THAP4
|
THAP domain containing 4 |
| chr1_-_245027833 | 0.53 |
ENST00000444376.2
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr18_+_9136758 | 0.53 |
ENST00000383440.2
ENST00000262126.4 ENST00000577992.1 |
ANKRD12
|
ankyrin repeat domain 12 |
| chr9_+_97136833 | 0.53 |
ENST00000375344.3
|
HIATL1
|
hippocampus abundant transcript-like 1 |
| chr17_-_42277203 | 0.52 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr6_-_52926539 | 0.51 |
ENST00000350082.5
ENST00000356971.3 |
ICK
|
intestinal cell (MAK-like) kinase |
| chr2_+_28113583 | 0.51 |
ENST00000344773.2
ENST00000379624.1 ENST00000342045.2 ENST00000379632.2 ENST00000361704.2 |
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr13_-_95364389 | 0.49 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr17_-_10101868 | 0.49 |
ENST00000432992.2
ENST00000540214.1 |
GAS7
|
growth arrest-specific 7 |
| chr18_-_44676819 | 0.49 |
ENST00000590815.1
ENST00000587388.1 ENST00000590481.1 ENST00000591480.1 ENST00000592591.1 ENST00000300605.6 |
HDHD2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr5_+_112312399 | 0.47 |
ENST00000515408.1
ENST00000513585.1 |
DCP2
|
decapping mRNA 2 |
| chr5_+_99871004 | 0.47 |
ENST00000312637.4
|
FAM174A
|
family with sequence similarity 174, member A |
| chr13_+_27131798 | 0.47 |
ENST00000361042.4
|
WASF3
|
WAS protein family, member 3 |
| chr9_+_91933407 | 0.47 |
ENST00000375807.3
ENST00000339901.4 |
SECISBP2
|
SECIS binding protein 2 |
| chr10_-_27529486 | 0.47 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr2_-_219906220 | 0.47 |
ENST00000458526.1
ENST00000409865.3 ENST00000410037.1 ENST00000457968.1 ENST00000436631.1 ENST00000341552.5 ENST00000441968.1 ENST00000295729.2 |
CCDC108
|
coiled-coil domain containing 108 |
| chr7_+_89841024 | 0.46 |
ENST00000394626.1
|
STEAP2
|
STEAP family member 2, metalloreductase |
| chr10_-_27529716 | 0.46 |
ENST00000375897.3
ENST00000396271.3 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr14_-_77495007 | 0.46 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr13_-_110438914 | 0.46 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr3_-_142682178 | 0.46 |
ENST00000340634.3
|
PAQR9
|
progestin and adipoQ receptor family member IX |
| chr4_-_54457783 | 0.45 |
ENST00000263925.7
ENST00000512247.1 |
LNX1
|
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| chr13_+_27131887 | 0.44 |
ENST00000335327.5
|
WASF3
|
WAS protein family, member 3 |
| chr12_-_88535842 | 0.44 |
ENST00000550962.1
ENST00000552810.1 |
CEP290
|
centrosomal protein 290kDa |
| chr15_+_51973550 | 0.44 |
ENST00000220478.3
|
SCG3
|
secretogranin III |
| chr16_-_28937027 | 0.43 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr2_-_38830030 | 0.43 |
ENST00000410076.1
|
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L-like |
| chrX_-_83442915 | 0.43 |
ENST00000262752.2
ENST00000543399.1 |
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
| chr2_+_46769798 | 0.42 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr1_+_183441500 | 0.42 |
ENST00000456731.2
|
SMG7
|
SMG7 nonsense mediated mRNA decay factor |
| chr19_-_48867291 | 0.42 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr7_+_89841000 | 0.42 |
ENST00000287908.3
|
STEAP2
|
STEAP family member 2, metalloreductase |
| chr10_-_119134918 | 0.42 |
ENST00000334464.5
|
PDZD8
|
PDZ domain containing 8 |
| chr19_-_56110859 | 0.42 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr11_-_32456891 | 0.42 |
ENST00000452863.3
|
WT1
|
Wilms tumor 1 |
| chrX_+_130192216 | 0.41 |
ENST00000276211.5
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr14_+_96968707 | 0.41 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr15_+_51973680 | 0.40 |
ENST00000542355.2
|
SCG3
|
secretogranin III |
| chr12_-_102224457 | 0.40 |
ENST00000549165.1
ENST00000549940.1 ENST00000392919.4 |
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr1_-_45805667 | 0.40 |
ENST00000488731.2
ENST00000435155.1 |
MUTYH
|
mutY homolog |
| chr15_-_77712477 | 0.40 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr15_+_45422131 | 0.40 |
ENST00000321429.4
|
DUOX1
|
dual oxidase 1 |
| chr22_+_41865109 | 0.39 |
ENST00000216254.4
ENST00000396512.3 |
ACO2
|
aconitase 2, mitochondrial |
| chr5_+_93954039 | 0.38 |
ENST00000265140.5
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr9_+_114393581 | 0.38 |
ENST00000313525.3
|
DNAJC25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr1_-_45805607 | 0.38 |
ENST00000372104.1
ENST00000448481.1 ENST00000483127.1 ENST00000528013.2 ENST00000456914.2 |
MUTYH
|
mutY homolog |
| chr6_+_126112001 | 0.38 |
ENST00000392477.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr1_+_235491714 | 0.38 |
ENST00000471812.1
ENST00000358966.2 ENST00000282841.5 ENST00000391855.2 |
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr15_+_45422178 | 0.38 |
ENST00000389037.3
ENST00000558322.1 |
DUOX1
|
dual oxidase 1 |
| chr9_+_140125385 | 0.38 |
ENST00000361134.2
|
SLC34A3
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr6_-_119670919 | 0.37 |
ENST00000368468.3
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1 |
| chr12_+_51632638 | 0.37 |
ENST00000549732.2
|
DAZAP2
|
DAZ associated protein 2 |
| chr15_-_83736091 | 0.37 |
ENST00000261721.4
|
BTBD1
|
BTB (POZ) domain containing 1 |
| chr12_+_69633317 | 0.36 |
ENST00000435070.2
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr1_-_74663825 | 0.36 |
ENST00000370911.3
ENST00000370909.2 ENST00000354431.4 |
LRRIQ3
|
leucine-rich repeats and IQ motif containing 3 |
| chr10_+_134901388 | 0.36 |
ENST00000392607.3
|
GPR123
|
G protein-coupled receptor 123 |
| chr12_+_51632508 | 0.36 |
ENST00000449723.3
|
DAZAP2
|
DAZ associated protein 2 |
| chr1_+_214454492 | 0.36 |
ENST00000366957.5
ENST00000415093.2 |
SMYD2
|
SET and MYND domain containing 2 |
| chr1_-_46152174 | 0.36 |
ENST00000290795.3
ENST00000355105.3 |
GPBP1L1
|
GC-rich promoter binding protein 1-like 1 |
| chr12_-_88535747 | 0.35 |
ENST00000309041.7
|
CEP290
|
centrosomal protein 290kDa |
| chr20_-_25062767 | 0.35 |
ENST00000429762.3
ENST00000444511.2 ENST00000376707.3 |
VSX1
|
visual system homeobox 1 |
| chr1_+_45805342 | 0.35 |
ENST00000372090.5
|
TOE1
|
target of EGR1, member 1 (nuclear) |
| chr5_+_110559603 | 0.34 |
ENST00000512453.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr14_-_96830207 | 0.34 |
ENST00000359933.4
|
ATG2B
|
autophagy related 2B |
| chr5_-_93447333 | 0.34 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr5_-_93954227 | 0.33 |
ENST00000513200.3
ENST00000329378.7 ENST00000312498.7 |
KIAA0825
|
KIAA0825 |
| chr2_+_170655789 | 0.33 |
ENST00000409333.1
|
SSB
|
Sjogren syndrome antigen B (autoantigen La) |
| chr6_-_150039249 | 0.33 |
ENST00000543571.1
|
LATS1
|
large tumor suppressor kinase 1 |
| chr8_+_141521386 | 0.32 |
ENST00000220913.5
ENST00000519533.1 |
CHRAC1
|
chromatin accessibility complex 1 |
| chr15_-_55700457 | 0.32 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr19_-_16770915 | 0.32 |
ENST00000593459.1
ENST00000358726.6 ENST00000597711.1 ENST00000487416.2 |
CTC-429P9.4
SMIM7
|
Small integral membrane protein 7; Uncharacterized protein small integral membrane protein 7 |
| chr19_-_48867171 | 0.32 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr3_+_137906109 | 0.32 |
ENST00000481646.1
ENST00000469044.1 ENST00000491704.1 ENST00000461600.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr1_-_53704157 | 0.32 |
ENST00000371466.4
ENST00000371470.3 |
MAGOH
|
mago-nashi homolog, proliferation-associated (Drosophila) |
| chr19_-_19144243 | 0.32 |
ENST00000594445.1
ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2
|
SURP and G patch domain containing 2 |
| chr1_-_235491462 | 0.31 |
ENST00000418304.1
ENST00000264183.3 ENST00000349213.3 |
ARID4B
|
AT rich interactive domain 4B (RBP1-like) |
| chr1_+_101361626 | 0.31 |
ENST00000370112.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr12_-_54071181 | 0.31 |
ENST00000338662.5
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr19_+_19030478 | 0.31 |
ENST00000247003.4
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr20_-_31172598 | 0.30 |
ENST00000201961.2
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr1_+_172502336 | 0.30 |
ENST00000263688.3
|
SUCO
|
SUN domain containing ossification factor |
| chr10_+_14880287 | 0.30 |
ENST00000437161.2
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr5_+_122847781 | 0.30 |
ENST00000395412.1
ENST00000395411.1 ENST00000345990.4 |
CSNK1G3
|
casein kinase 1, gamma 3 |
| chr2_-_73520667 | 0.30 |
ENST00000545030.1
ENST00000436467.2 |
EGR4
|
early growth response 4 |
| chr10_+_104005272 | 0.30 |
ENST00000369983.3
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr7_+_131012605 | 0.30 |
ENST00000446815.1
ENST00000352689.6 |
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr8_-_38126635 | 0.29 |
ENST00000529359.1
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr6_+_126112074 | 0.29 |
ENST00000453302.1
ENST00000417494.1 ENST00000229634.9 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr17_-_33905521 | 0.29 |
ENST00000225873.4
|
PEX12
|
peroxisomal biogenesis factor 12 |
| chr18_-_19180681 | 0.29 |
ENST00000269214.5
|
ESCO1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr1_-_28241024 | 0.29 |
ENST00000313433.7
ENST00000444045.1 |
RPA2
|
replication protein A2, 32kDa |
| chr19_+_19144384 | 0.28 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr9_+_33025209 | 0.28 |
ENST00000330899.4
ENST00000544625.1 |
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr15_+_49447947 | 0.28 |
ENST00000327171.3
ENST00000560654.1 |
GALK2
|
galactokinase 2 |
| chr10_-_125851961 | 0.28 |
ENST00000346248.5
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr8_-_38126675 | 0.26 |
ENST00000531823.1
ENST00000534339.1 ENST00000524616.1 ENST00000422581.2 ENST00000424479.2 ENST00000419686.2 |
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr7_+_92158083 | 0.26 |
ENST00000265732.5
ENST00000481551.1 ENST00000496410.1 |
RBM48
|
RNA binding motif protein 48 |
| chr5_+_32585605 | 0.26 |
ENST00000265073.4
ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr14_+_92588281 | 0.26 |
ENST00000298875.4
ENST00000553427.1 |
CPSF2
|
cleavage and polyadenylation specific factor 2, 100kDa |
| chr12_+_56110315 | 0.26 |
ENST00000548556.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chrX_-_99891796 | 0.26 |
ENST00000373020.4
|
TSPAN6
|
tetraspanin 6 |
| chr6_+_27215494 | 0.25 |
ENST00000230582.3
|
PRSS16
|
protease, serine, 16 (thymus) |
| chr8_+_125551338 | 0.25 |
ENST00000276689.3
ENST00000518008.1 ENST00000522532.1 ENST00000517367.1 |
NDUFB9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa |
| chr12_+_51632600 | 0.25 |
ENST00000549555.1
ENST00000439799.2 ENST00000425012.2 |
DAZAP2
|
DAZ associated protein 2 |
| chr11_-_61684962 | 0.25 |
ENST00000394836.2
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr12_+_56110374 | 0.25 |
ENST00000549424.1
|
RP11-644F5.10
|
Uncharacterized protein |
| chr19_-_10444188 | 0.25 |
ENST00000293677.6
|
RAVER1
|
ribonucleoprotein, PTB-binding 1 |
| chr17_+_11501816 | 0.24 |
ENST00000454412.2
|
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr6_+_27215471 | 0.24 |
ENST00000421826.2
|
PRSS16
|
protease, serine, 16 (thymus) |
| chr6_+_30457244 | 0.24 |
ENST00000376630.4
|
HLA-E
|
major histocompatibility complex, class I, E |
| chr16_-_75498553 | 0.24 |
ENST00000569276.1
ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A
RP11-77K12.1
|
transmembrane protein 170A Uncharacterized protein |
| chr19_-_37663332 | 0.24 |
ENST00000392157.2
|
ZNF585A
|
zinc finger protein 585A |
| chr2_-_38829768 | 0.24 |
ENST00000378915.3
|
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr17_-_26972126 | 0.23 |
ENST00000528896.2
|
KIAA0100
|
KIAA0100 |
| chr17_+_7387919 | 0.23 |
ENST00000572844.1
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr12_+_120933904 | 0.23 |
ENST00000550178.1
ENST00000550845.1 ENST00000549989.1 ENST00000552870.1 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr12_+_56109926 | 0.22 |
ENST00000547076.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr4_+_48343339 | 0.22 |
ENST00000264313.6
|
SLAIN2
|
SLAIN motif family, member 2 |
| chr11_+_61447845 | 0.22 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr10_-_127408011 | 0.22 |
ENST00000531977.1
ENST00000527483.1 ENST00000525909.1 ENST00000528844.1 ENST00000423178.2 |
RP11-383C5.4
|
RP11-383C5.4 |
| chr2_-_20251744 | 0.22 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chrX_-_71525742 | 0.21 |
ENST00000450875.1
ENST00000417400.1 ENST00000431381.1 ENST00000445983.1 |
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr12_+_49740700 | 0.21 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr11_-_82782952 | 0.21 |
ENST00000534141.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr15_+_41221536 | 0.21 |
ENST00000249749.5
|
DLL4
|
delta-like 4 (Drosophila) |
| chr1_-_28241226 | 0.21 |
ENST00000373912.3
ENST00000373909.3 |
RPA2
|
replication protein A2, 32kDa |
| chr2_+_54198210 | 0.21 |
ENST00000607452.1
ENST00000422521.2 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr7_-_92157747 | 0.20 |
ENST00000428214.1
ENST00000438045.1 |
PEX1
|
peroxisomal biogenesis factor 1 |
| chr14_+_96968802 | 0.20 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXN1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.4 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.4 | 1.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.3 | 1.9 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.2 | 0.7 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.2 | 3.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 2.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 0.6 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 1.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.6 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.4 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 2.0 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.8 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.9 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.9 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.1 | 2.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.0 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.1 | 0.8 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 1.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.9 | GO:0033216 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 1.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.4 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.1 | 0.4 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.8 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.7 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.7 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.5 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 0.3 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.4 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.8 | GO:0030916 | otic vesicle formation(GO:0030916) pronephros development(GO:0048793) |
| 0.1 | 0.4 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.1 | 0.2 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.1 | 1.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.2 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.1 | 0.5 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 2.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 0.5 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 1.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.5 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 1.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 2.2 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.9 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.7 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.3 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.9 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.5 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.4 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.1 | GO:0043634 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.0 | 0.6 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.4 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.0 | 1.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 1.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 2.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.4 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.3 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 1.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.0 | GO:0035607 | vacuolar phosphate transport(GO:0007037) fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development(GO:0035607) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.3 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.0 | 0.2 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.2 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 1.3 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 0.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 4.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 1.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 2.3 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 2.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.5 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 3.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) interchromatin granule(GO:0035061) |
| 0.0 | 1.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 1.9 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.3 | 0.9 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.3 | 1.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 0.8 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.2 | 6.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 0.9 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.7 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.4 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 4.7 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.7 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.4 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 1.6 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.9 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.8 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.3 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 0.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.4 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.5 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.5 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 2.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 1.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.6 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 1.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 1.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 3.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.1 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.8 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 2.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 2.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.8 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |