Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
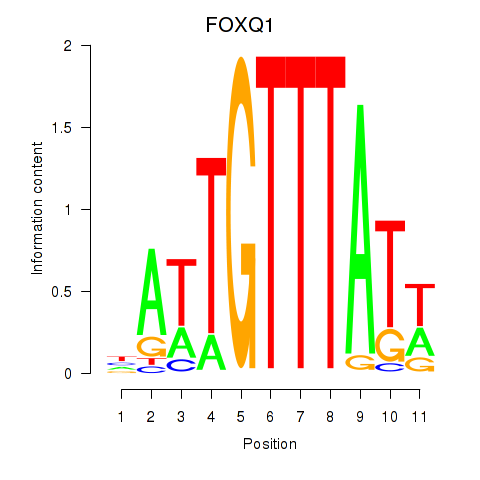
Results for FOXQ1
Z-value: 0.71
Transcription factors associated with FOXQ1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXQ1
|
ENSG00000164379.4 | forkhead box Q1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXQ1 | hg19_v2_chr6_+_1312675_1312701 | 0.36 | 5.4e-02 | Click! |
Activity profile of FOXQ1 motif
Sorted Z-values of FOXQ1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_94727048 | 2.99 |
ENST00000283357.5
|
FAM81B
|
family with sequence similarity 81, member B |
| chr12_-_71551652 | 2.94 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr4_+_165675269 | 2.69 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr6_+_131958436 | 2.45 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr13_-_39564993 | 2.36 |
ENST00000423210.1
|
STOML3
|
stomatin (EPB72)-like 3 |
| chr3_-_19975665 | 2.26 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chr3_-_167371740 | 2.16 |
ENST00000466760.1
ENST00000479765.1 |
WDR49
|
WD repeat domain 49 |
| chr9_+_124922171 | 2.05 |
ENST00000373764.3
ENST00000536616.1 |
MORN5
|
MORN repeat containing 5 |
| chr14_+_96949319 | 1.99 |
ENST00000554706.1
|
AK7
|
adenylate kinase 7 |
| chr4_+_165675197 | 1.94 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr6_-_52774464 | 1.89 |
ENST00000370968.1
ENST00000211122.3 |
GSTA3
|
glutathione S-transferase alpha 3 |
| chr11_+_86106208 | 1.88 |
ENST00000528728.1
|
CCDC81
|
coiled-coil domain containing 81 |
| chr5_-_13944652 | 1.79 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr11_+_71791359 | 1.74 |
ENST00000419228.1
ENST00000435085.1 ENST00000307198.7 ENST00000538413.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr1_+_104159999 | 1.68 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chrY_+_15016013 | 1.62 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr6_+_123038689 | 1.61 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr11_+_6897856 | 1.60 |
ENST00000379829.2
|
OR10A4
|
olfactory receptor, family 10, subfamily A, member 4 |
| chr12_-_71551868 | 1.54 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr15_-_56757329 | 1.44 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr1_-_216978709 | 1.37 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr5_+_121465207 | 1.29 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr11_+_60467047 | 1.27 |
ENST00000300226.2
|
MS4A8
|
membrane-spanning 4-domains, subfamily A, member 8 |
| chr8_+_94767109 | 1.24 |
ENST00000409623.3
ENST00000453906.1 ENST00000521517.1 |
TMEM67
|
transmembrane protein 67 |
| chr10_+_134150835 | 1.23 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr11_-_26593779 | 1.18 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr9_-_35563896 | 1.16 |
ENST00000399742.2
|
FAM166B
|
family with sequence similarity 166, member B |
| chr19_-_48823332 | 1.13 |
ENST00000315396.7
|
CCDC114
|
coiled-coil domain containing 114 |
| chr11_-_26593677 | 1.07 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr8_+_94767072 | 1.07 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr16_+_4784273 | 1.06 |
ENST00000299320.5
ENST00000586724.1 |
C16orf71
|
chromosome 16 open reading frame 71 |
| chr11_-_26593649 | 1.02 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr6_-_32731299 | 1.01 |
ENST00000435145.2
ENST00000437316.2 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr10_+_127661942 | 0.97 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr10_-_75118611 | 0.88 |
ENST00000355577.3
ENST00000394865.1 ENST00000310715.3 ENST00000401621.2 |
TTC18
|
tetratricopeptide repeat domain 18 |
| chr19_-_14992264 | 0.88 |
ENST00000327462.2
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17 |
| chr16_+_4784458 | 0.84 |
ENST00000590191.1
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr6_-_52668605 | 0.84 |
ENST00000334575.5
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr19_+_709101 | 0.82 |
ENST00000338448.5
|
PALM
|
paralemmin |
| chr4_-_186696425 | 0.80 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr17_+_43238061 | 0.78 |
ENST00000307275.3
ENST00000585340.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr1_-_54405773 | 0.77 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr3_-_112564797 | 0.76 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr14_-_23288930 | 0.76 |
ENST00000554517.1
ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr6_-_9939552 | 0.76 |
ENST00000460363.2
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr3_+_171561127 | 0.72 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr7_-_122635754 | 0.72 |
ENST00000249284.2
|
TAS2R16
|
taste receptor, type 2, member 16 |
| chr5_+_140571902 | 0.71 |
ENST00000239446.4
|
PCDHB10
|
protocadherin beta 10 |
| chr15_+_50474385 | 0.70 |
ENST00000267842.5
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2 |
| chr7_-_130080977 | 0.70 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr1_+_171060018 | 0.70 |
ENST00000367755.4
ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3
|
flavin containing monooxygenase 3 |
| chr1_+_63989004 | 0.67 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr1_+_61330931 | 0.66 |
ENST00000371191.1
|
NFIA
|
nuclear factor I/A |
| chr21_-_31588365 | 0.64 |
ENST00000399899.1
|
CLDN8
|
claudin 8 |
| chr5_-_41261540 | 0.62 |
ENST00000263413.3
|
C6
|
complement component 6 |
| chr3_+_190333097 | 0.62 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr3_-_57530051 | 0.61 |
ENST00000311202.6
ENST00000351747.2 ENST00000495027.1 ENST00000389536.4 |
DNAH12
|
dynein, axonemal, heavy chain 12 |
| chr12_-_49582978 | 0.60 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr1_+_61542922 | 0.60 |
ENST00000407417.3
|
NFIA
|
nuclear factor I/A |
| chr21_-_31588338 | 0.60 |
ENST00000286809.1
|
CLDN8
|
claudin 8 |
| chr1_-_227505289 | 0.59 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr5_+_73109339 | 0.58 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr14_+_21498360 | 0.56 |
ENST00000321760.6
ENST00000460647.2 ENST00000530140.2 ENST00000472458.1 |
TPPP2
|
tubulin polymerization-promoting protein family member 2 |
| chrX_-_108725301 | 0.56 |
ENST00000218006.2
|
GUCY2F
|
guanylate cyclase 2F, retinal |
| chr16_+_30212378 | 0.56 |
ENST00000569485.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr1_+_174844645 | 0.55 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr7_+_48211048 | 0.55 |
ENST00000435803.1
|
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr11_-_1629693 | 0.55 |
ENST00000399685.1
|
KRTAP5-3
|
keratin associated protein 5-3 |
| chr15_+_50474412 | 0.55 |
ENST00000380902.4
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2 |
| chr2_-_157198860 | 0.55 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr6_-_52710893 | 0.53 |
ENST00000284562.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr14_-_89878369 | 0.53 |
ENST00000553840.1
ENST00000556916.1 |
FOXN3
|
forkhead box N3 |
| chr12_+_19358228 | 0.52 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr3_+_108321623 | 0.51 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr9_+_2029019 | 0.51 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_+_21168630 | 0.51 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr2_-_55459294 | 0.51 |
ENST00000407122.1
ENST00000406437.2 |
CLHC1
|
clathrin heavy chain linker domain containing 1 |
| chr3_-_114477962 | 0.50 |
ENST00000471418.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr15_+_49715293 | 0.49 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr3_-_114343039 | 0.49 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_+_114472222 | 0.48 |
ENST00000369558.1
ENST00000369561.4 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr18_+_77160282 | 0.47 |
ENST00000318065.5
ENST00000545796.1 ENST00000592223.1 ENST00000329101.4 ENST00000586434.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr12_+_1738363 | 0.47 |
ENST00000397196.2
|
WNT5B
|
wingless-type MMTV integration site family, member 5B |
| chr4_+_40751914 | 0.46 |
ENST00000381782.2
ENST00000316607.5 |
NSUN7
|
NOP2/Sun domain family, member 7 |
| chr17_+_73642315 | 0.45 |
ENST00000556126.2
|
SMIM6
|
small integral membrane protein 6 |
| chr1_-_149832704 | 0.44 |
ENST00000392933.1
ENST00000369157.2 ENST00000392932.4 |
HIST2H4B
|
histone cluster 2, H4b |
| chr3_-_151102529 | 0.44 |
ENST00000302632.3
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12 |
| chr9_-_126030817 | 0.44 |
ENST00000348403.5
ENST00000447404.2 ENST00000360998.3 |
STRBP
|
spermatid perinuclear RNA binding protein |
| chr6_-_87804815 | 0.44 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr10_+_101542462 | 0.43 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr6_-_31648150 | 0.43 |
ENST00000375858.3
ENST00000383237.4 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr1_+_70820451 | 0.43 |
ENST00000361764.4
ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3
|
HERV-H LTR-associating 3 |
| chr6_-_32731243 | 0.42 |
ENST00000427449.1
ENST00000411527.1 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr12_+_74931551 | 0.42 |
ENST00000519948.2
|
ATXN7L3B
|
ataxin 7-like 3B |
| chr1_-_76398077 | 0.41 |
ENST00000284142.6
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr6_-_49681235 | 0.41 |
ENST00000339139.4
|
CRISP2
|
cysteine-rich secretory protein 2 |
| chr17_+_72426891 | 0.41 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr10_-_49482907 | 0.40 |
ENST00000374201.3
ENST00000407470.4 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr11_-_85397167 | 0.39 |
ENST00000316398.3
|
CCDC89
|
coiled-coil domain containing 89 |
| chr6_-_168479839 | 0.39 |
ENST00000283309.6
|
FRMD1
|
FERM domain containing 1 |
| chr5_+_140227048 | 0.38 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr11_+_121447469 | 0.38 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr14_+_31046959 | 0.38 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr9_-_13279589 | 0.37 |
ENST00000319217.7
|
MPDZ
|
multiple PDZ domain protein |
| chr11_+_10476851 | 0.37 |
ENST00000396553.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr5_+_140557371 | 0.37 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr9_-_13279563 | 0.37 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr14_+_102276132 | 0.36 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr1_+_114471809 | 0.36 |
ENST00000426820.2
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr4_-_119274121 | 0.36 |
ENST00000296498.3
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr7_-_64023441 | 0.36 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr1_-_150669500 | 0.36 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr18_-_57027194 | 0.35 |
ENST00000251047.5
|
LMAN1
|
lectin, mannose-binding, 1 |
| chr12_-_122879969 | 0.35 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr6_+_140175987 | 0.35 |
ENST00000414038.1
ENST00000431609.1 |
RP5-899B16.1
|
RP5-899B16.1 |
| chr1_+_236686717 | 0.34 |
ENST00000341872.6
ENST00000450372.2 |
LGALS8
|
lectin, galactoside-binding, soluble, 8 |
| chr7_-_122342966 | 0.34 |
ENST00000447240.1
|
RNF148
|
ring finger protein 148 |
| chr7_-_122342988 | 0.34 |
ENST00000434824.1
|
RNF148
|
ring finger protein 148 |
| chr9_+_78505581 | 0.34 |
ENST00000376767.3
ENST00000376752.4 |
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr8_+_110551925 | 0.33 |
ENST00000395785.2
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr18_-_53257027 | 0.33 |
ENST00000568740.1
ENST00000564403.2 ENST00000537578.1 |
TCF4
|
transcription factor 4 |
| chr18_+_5238055 | 0.33 |
ENST00000582363.1
ENST00000582008.1 ENST00000580082.1 |
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr5_+_140625147 | 0.32 |
ENST00000231173.3
|
PCDHB15
|
protocadherin beta 15 |
| chr4_+_111397216 | 0.31 |
ENST00000265162.5
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A) |
| chr4_+_146539415 | 0.31 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr8_+_79428539 | 0.31 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr7_+_54610124 | 0.31 |
ENST00000402026.2
|
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chrX_+_49020121 | 0.31 |
ENST00000415364.1
ENST00000376338.3 ENST00000425285.1 |
MAGIX
|
MAGI family member, X-linked |
| chr12_+_109554386 | 0.31 |
ENST00000338432.7
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr9_+_78505554 | 0.31 |
ENST00000545128.1
|
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr6_-_52705641 | 0.30 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr10_+_11784360 | 0.30 |
ENST00000379215.4
ENST00000420401.1 |
ECHDC3
|
enoyl CoA hydratase domain containing 3 |
| chr18_-_21852143 | 0.30 |
ENST00000399443.3
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr9_-_15472730 | 0.30 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr6_-_29013017 | 0.29 |
ENST00000377175.1
|
OR2W1
|
olfactory receptor, family 2, subfamily W, member 1 |
| chr14_+_105267250 | 0.29 |
ENST00000342537.7
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr3_+_35721182 | 0.29 |
ENST00000413378.1
ENST00000417925.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr6_-_24666819 | 0.29 |
ENST00000341060.3
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr9_-_123239632 | 0.29 |
ENST00000416449.1
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chrX_+_49020882 | 0.28 |
ENST00000454342.1
|
MAGIX
|
MAGI family member, X-linked |
| chr14_+_50999744 | 0.28 |
ENST00000441560.2
|
ATL1
|
atlastin GTPase 1 |
| chr13_+_103459704 | 0.28 |
ENST00000602836.1
|
BIVM-ERCC5
|
BIVM-ERCC5 readthrough |
| chr11_+_112047087 | 0.27 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr2_+_163175394 | 0.27 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr5_+_140220769 | 0.27 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chr14_-_34420259 | 0.27 |
ENST00000250457.3
ENST00000547327.2 |
EGLN3
|
egl-9 family hypoxia-inducible factor 3 |
| chr5_+_93954039 | 0.27 |
ENST00000265140.5
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr12_+_121570631 | 0.26 |
ENST00000546057.1
ENST00000377162.2 ENST00000328963.5 ENST00000535250.1 ENST00000541446.1 |
P2RX7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chrX_+_105855160 | 0.26 |
ENST00000372544.2
ENST00000372548.4 |
CXorf57
|
chromosome X open reading frame 57 |
| chr14_+_51026743 | 0.26 |
ENST00000358385.6
ENST00000357032.3 ENST00000354525.4 |
ATL1
|
atlastin GTPase 1 |
| chr2_+_47596287 | 0.26 |
ENST00000263735.4
|
EPCAM
|
epithelial cell adhesion molecule |
| chr6_-_136871957 | 0.26 |
ENST00000354570.3
|
MAP7
|
microtubule-associated protein 7 |
| chr4_+_17578815 | 0.26 |
ENST00000226299.4
|
LAP3
|
leucine aminopeptidase 3 |
| chr6_+_26501449 | 0.26 |
ENST00000244513.6
|
BTN1A1
|
butyrophilin, subfamily 1, member A1 |
| chr3_+_138340049 | 0.26 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr21_+_40824003 | 0.25 |
ENST00000452550.1
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein |
| chr2_+_214149113 | 0.25 |
ENST00000331683.5
ENST00000432529.2 ENST00000413312.1 ENST00000272898.7 ENST00000447990.1 |
SPAG16
|
sperm associated antigen 16 |
| chr5_+_64920543 | 0.25 |
ENST00000399438.3
ENST00000510585.2 |
TRAPPC13
CTC-534A2.2
|
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr8_+_110552046 | 0.25 |
ENST00000529931.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr5_-_93954227 | 0.25 |
ENST00000513200.3
ENST00000329378.7 ENST00000312498.7 |
KIAA0825
|
KIAA0825 |
| chr11_+_22688150 | 0.25 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr11_-_115375107 | 0.24 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr5_+_42423872 | 0.24 |
ENST00000230882.4
ENST00000357703.3 |
GHR
|
growth hormone receptor |
| chr5_+_118690466 | 0.24 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr21_+_25801041 | 0.24 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr12_+_72233487 | 0.24 |
ENST00000482439.2
ENST00000550746.1 ENST00000491063.1 ENST00000319106.8 ENST00000485960.2 ENST00000393309.3 |
TBC1D15
|
TBC1 domain family, member 15 |
| chr5_+_72921983 | 0.24 |
ENST00000296794.6
ENST00000545377.1 ENST00000513042.2 ENST00000287898.5 ENST00000509848.1 |
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr2_-_165424973 | 0.24 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr1_-_179112189 | 0.24 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr17_+_79953310 | 0.24 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr11_-_107328527 | 0.23 |
ENST00000282251.5
ENST00000433523.1 |
CWF19L2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr2_-_203735976 | 0.23 |
ENST00000435143.1
|
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr6_-_28554977 | 0.23 |
ENST00000452236.2
|
SCAND3
|
SCAN domain containing 3 |
| chr8_+_109455845 | 0.23 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr17_+_19186292 | 0.23 |
ENST00000395626.1
ENST00000571254.1 |
EPN2
|
epsin 2 |
| chr6_+_53883790 | 0.23 |
ENST00000509997.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr16_+_69958887 | 0.23 |
ENST00000568684.1
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr4_-_120225600 | 0.23 |
ENST00000399075.4
|
C4orf3
|
chromosome 4 open reading frame 3 |
| chr17_-_19648683 | 0.22 |
ENST00000573368.1
ENST00000457500.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr1_+_239882842 | 0.22 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr3_+_157154578 | 0.22 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr1_+_10292308 | 0.22 |
ENST00000377081.1
|
KIF1B
|
kinesin family member 1B |
| chr10_-_98031265 | 0.22 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr6_-_52628271 | 0.21 |
ENST00000493422.1
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr5_-_111091948 | 0.21 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr4_+_146403912 | 0.21 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr2_+_211342432 | 0.21 |
ENST00000430249.2
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr16_+_28763108 | 0.21 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr5_-_218251 | 0.21 |
ENST00000296824.3
|
CCDC127
|
coiled-coil domain containing 127 |
| chr16_-_15474904 | 0.21 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr11_+_58710722 | 0.21 |
ENST00000300079.5
|
GLYATL1
|
glycine-N-acyltransferase-like 1 |
| chr11_-_102668879 | 0.20 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr14_+_65381079 | 0.20 |
ENST00000549115.1
ENST00000607599.1 ENST00000548752.2 ENST00000359118.2 ENST00000552002.2 ENST00000551947.1 ENST00000551093.1 ENST00000542227.1 ENST00000447296.2 ENST00000549987.1 |
CHURC1
FNTB
CHURC1-FNTB
|
churchill domain containing 1 farnesyltransferase, CAAX box, beta CHURC1-FNTB readthrough |
| chr8_-_124553437 | 0.20 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr16_+_14802801 | 0.20 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr3_+_138340067 | 0.20 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr9_+_4662282 | 0.19 |
ENST00000381883.2
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2 |
| chr11_-_8285405 | 0.19 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr12_-_18243119 | 0.19 |
ENST00000538724.1
ENST00000229002.2 |
RERGL
|
RERG/RAS-like |
| chr9_-_85882145 | 0.19 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXQ1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.3 | 1.3 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.2 | 1.7 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.2 | 0.5 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.2 | 0.6 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.1 | 0.4 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.1 | 2.6 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.1 | 2.9 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.6 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 0.4 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.8 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 2.3 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 2.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.3 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 0.1 | 0.6 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.8 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 0.8 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.3 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.1 | 1.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 0.3 | GO:1904170 | NAD transport(GO:0043132) regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.1 | 0.5 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.1 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.1 | 0.3 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.3 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 2.9 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 0.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.3 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.1 | 0.4 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.4 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.2 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 0.2 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.1 | 0.2 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.2 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.2 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.1 | 0.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.3 | GO:0044254 | angiotensin catabolic process in blood(GO:0002005) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.2 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 3.7 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.3 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 1.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.0 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.2 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 2.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 2.0 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0043474 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) regulation of melanosome transport(GO:1902908) |
| 0.0 | 0.8 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 1.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 1.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0044854 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.0 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.4 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.1 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.0 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 2.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.3 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 0.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 2.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.5 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 1.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.4 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 1.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 3.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.4 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.0 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 1.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.4 | 1.7 | GO:0016160 | amylase activity(GO:0016160) |
| 0.3 | 2.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.2 | 1.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 1.7 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.2 | 0.8 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.1 | 0.6 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 1.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.3 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.1 | 0.6 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 2.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 1.8 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.2 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 3.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.5 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 1.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.5 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 3.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 3.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 3.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 3.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |