Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for GATA1_GATA4
Z-value: 0.89
Transcription factors associated with GATA1_GATA4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
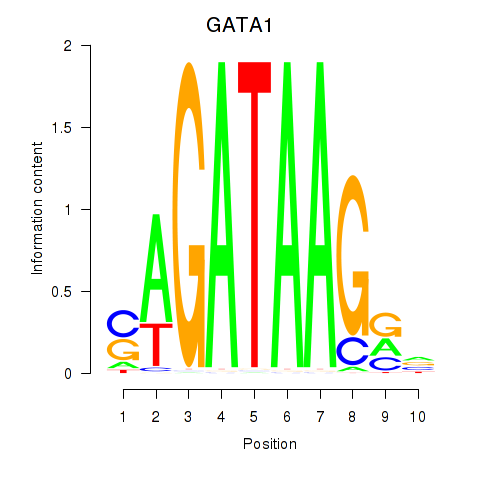
GATA1
|
ENSG00000102145.9 | GATA binding protein 1 |
|
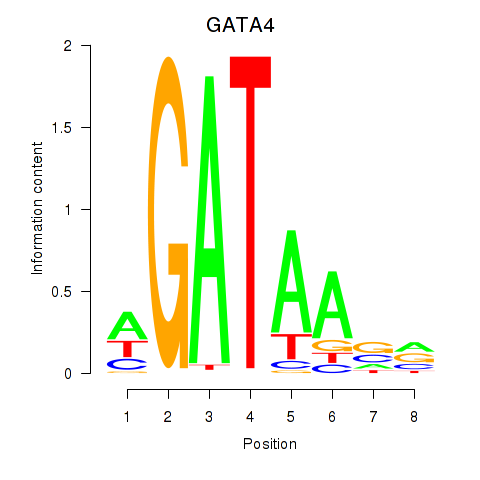
GATA4
|
ENSG00000136574.13 | GATA binding protein 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA1 | hg19_v2_chrX_+_48644962_48644983 | 0.55 | 1.5e-03 | Click! |
| GATA4 | hg19_v2_chr8_+_11534462_11534475 | 0.50 | 4.9e-03 | Click! |
Activity profile of GATA1_GATA4 motif
Sorted Z-values of GATA1_GATA4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_102928009 | 4.91 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr1_-_153113927 | 4.87 |
ENST00000368752.4
|
SPRR2B
|
small proline-rich protein 2B |
| chr9_+_33795533 | 4.69 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr1_-_24469602 | 4.23 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr12_+_4385230 | 3.86 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chr4_-_159094194 | 3.30 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr19_-_51487071 | 2.60 |
ENST00000391807.1
ENST00000593904.1 |
KLK7
|
kallikrein-related peptidase 7 |
| chr4_-_103266355 | 2.58 |
ENST00000424970.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr19_-_51487282 | 2.45 |
ENST00000595820.1
ENST00000597707.1 ENST00000336317.4 |
KLK7
|
kallikrein-related peptidase 7 |
| chrX_+_135278908 | 2.18 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr17_+_74381343 | 2.18 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr8_+_124194875 | 2.11 |
ENST00000522648.1
ENST00000276699.6 |
FAM83A
|
family with sequence similarity 83, member A |
| chr8_+_124194752 | 2.03 |
ENST00000318462.6
|
FAM83A
|
family with sequence similarity 83, member A |
| chr1_-_153029980 | 1.93 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr3_-_98312548 | 1.90 |
ENST00000264193.2
|
CPOX
|
coproporphyrinogen oxidase |
| chr1_-_153013588 | 1.85 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chrX_+_135279179 | 1.82 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr3_-_47950745 | 1.66 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr12_+_15699286 | 1.58 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chrX_+_56258844 | 1.54 |
ENST00000374928.3
|
KLF8
|
Kruppel-like factor 8 |
| chr2_-_165424973 | 1.53 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr7_-_99277610 | 1.52 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr9_+_139557360 | 1.47 |
ENST00000308874.7
ENST00000406555.3 ENST00000492862.2 |
EGFL7
|
EGF-like-domain, multiple 7 |
| chr4_-_103266219 | 1.46 |
ENST00000394833.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr7_+_100547156 | 1.46 |
ENST00000379458.4
|
MUC3A
|
Protein LOC100131514 |
| chr19_+_10397621 | 1.45 |
ENST00000380770.3
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr9_+_34652164 | 1.43 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr19_+_10397648 | 1.41 |
ENST00000340992.4
ENST00000393717.2 |
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr2_+_174219548 | 1.40 |
ENST00000347703.3
ENST00000392567.2 ENST00000306721.3 ENST00000410101.3 ENST00000410019.3 |
CDCA7
|
cell division cycle associated 7 |
| chr18_+_19749386 | 1.40 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr2_-_24307162 | 1.38 |
ENST00000413037.1
ENST00000407482.1 |
TP53I3
|
tumor protein p53 inducible protein 3 |
| chr16_+_70680439 | 1.37 |
ENST00000288098.2
|
IL34
|
interleukin 34 |
| chr9_-_114937465 | 1.37 |
ENST00000355396.3
|
SUSD1
|
sushi domain containing 1 |
| chr4_-_68749745 | 1.33 |
ENST00000283916.6
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chrX_+_152082969 | 1.32 |
ENST00000535861.1
ENST00000539731.1 ENST00000449285.2 ENST00000318504.7 ENST00000324823.6 ENST00000370268.4 ENST00000370270.2 |
ZNF185
|
zinc finger protein 185 (LIM domain) |
| chr5_+_159848807 | 1.27 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr9_-_114937543 | 1.27 |
ENST00000374264.2
ENST00000374263.3 |
SUSD1
|
sushi domain containing 1 |
| chr5_-_39219705 | 1.26 |
ENST00000351578.6
|
FYB
|
FYN binding protein |
| chr1_-_94050668 | 1.26 |
ENST00000539242.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr22_-_20231207 | 1.22 |
ENST00000425986.1
|
RTN4R
|
reticulon 4 receptor |
| chr6_+_12290586 | 1.22 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr9_+_33240157 | 1.20 |
ENST00000379721.3
|
SPINK4
|
serine peptidase inhibitor, Kazal type 4 |
| chr12_-_52845910 | 1.19 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chr1_-_153521597 | 1.18 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr17_-_73840415 | 1.18 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr7_-_50860565 | 1.18 |
ENST00000403097.1
|
GRB10
|
growth factor receptor-bound protein 10 |
| chr15_-_34610962 | 1.16 |
ENST00000290209.5
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr14_+_85996471 | 1.15 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chrX_+_107288239 | 1.15 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr17_+_33914276 | 1.13 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chrX_+_107288197 | 1.12 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr1_-_201368653 | 1.11 |
ENST00000367313.3
|
LAD1
|
ladinin 1 |
| chr2_+_27719697 | 1.10 |
ENST00000264717.2
ENST00000424318.2 |
GCKR
|
glucokinase (hexokinase 4) regulator |
| chr2_-_69098566 | 1.09 |
ENST00000295379.1
|
BMP10
|
bone morphogenetic protein 10 |
| chr9_-_114937676 | 1.08 |
ENST00000374270.3
|
SUSD1
|
sushi domain containing 1 |
| chr4_-_68749699 | 1.06 |
ENST00000545541.1
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr5_-_39270725 | 1.05 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr1_-_201368707 | 1.05 |
ENST00000391967.2
|
LAD1
|
ladinin 1 |
| chr5_-_39219641 | 1.03 |
ENST00000509072.1
ENST00000504542.1 ENST00000505428.1 ENST00000506557.1 |
FYB
|
FYN binding protein |
| chrX_+_44703249 | 1.02 |
ENST00000339042.4
|
DUSP21
|
dual specificity phosphatase 21 |
| chr9_+_132099158 | 1.01 |
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr7_+_142457315 | 1.01 |
ENST00000486171.1
ENST00000311737.7 |
PRSS1
|
protease, serine, 1 (trypsin 1) |
| chr1_-_204116078 | 1.01 |
ENST00000367198.2
ENST00000452983.1 |
ETNK2
|
ethanolamine kinase 2 |
| chrX_+_38211777 | 1.00 |
ENST00000039007.4
|
OTC
|
ornithine carbamoyltransferase |
| chr12_+_4430371 | 0.99 |
ENST00000179259.4
|
C12orf5
|
chromosome 12 open reading frame 5 |
| chr1_-_153521714 | 0.97 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr3_+_172468505 | 0.94 |
ENST00000427830.1
ENST00000417960.1 ENST00000428567.1 ENST00000366090.2 ENST00000426894.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr10_+_71075552 | 0.92 |
ENST00000298649.3
|
HK1
|
hexokinase 1 |
| chr6_-_131277510 | 0.92 |
ENST00000525193.1
ENST00000527659.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr14_-_94421923 | 0.92 |
ENST00000555507.1
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chr4_-_120243545 | 0.91 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr18_+_61445007 | 0.91 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr6_-_107235287 | 0.90 |
ENST00000436659.1
ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr8_-_91095099 | 0.90 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr17_+_1958388 | 0.90 |
ENST00000399849.3
|
HIC1
|
hypermethylated in cancer 1 |
| chr11_-_125550764 | 0.90 |
ENST00000527795.1
|
ACRV1
|
acrosomal vesicle protein 1 |
| chr5_+_147443534 | 0.89 |
ENST00000398454.1
ENST00000359874.3 ENST00000508733.1 ENST00000256084.7 |
SPINK5
|
serine peptidase inhibitor, Kazal type 5 |
| chr2_-_228028829 | 0.87 |
ENST00000396625.3
ENST00000329662.7 |
COL4A4
|
collagen, type IV, alpha 4 |
| chr2_+_192543153 | 0.86 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr7_+_139528952 | 0.86 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr17_+_19314505 | 0.85 |
ENST00000461366.1
|
RNF112
|
ring finger protein 112 |
| chr7_-_107443652 | 0.83 |
ENST00000340010.5
ENST00000422236.2 ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 (anion exchanger), member 3 |
| chrX_+_99899180 | 0.83 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr11_-_3859089 | 0.82 |
ENST00000396979.1
|
RHOG
|
ras homolog family member G |
| chr6_+_47666275 | 0.80 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr2_+_201994569 | 0.79 |
ENST00000457277.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr15_-_34628951 | 0.79 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr19_-_51522955 | 0.79 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chrX_-_65253506 | 0.78 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr15_-_74501360 | 0.78 |
ENST00000323940.5
|
STRA6
|
stimulated by retinoic acid 6 |
| chr8_+_70404996 | 0.78 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr1_+_25598872 | 0.78 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
| chr15_+_42066740 | 0.78 |
ENST00000514566.1
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr18_+_21452804 | 0.78 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr12_+_9144626 | 0.73 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr11_-_124311054 | 0.73 |
ENST00000328064.2
|
OR8B8
|
olfactory receptor, family 8, subfamily B, member 8 |
| chr2_+_201994208 | 0.73 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr11_-_125550726 | 0.72 |
ENST00000315608.3
ENST00000530048.1 |
ACRV1
|
acrosomal vesicle protein 1 |
| chr7_-_23510086 | 0.71 |
ENST00000258729.3
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr19_-_51568324 | 0.71 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr10_+_5566916 | 0.70 |
ENST00000315238.1
|
CALML3
|
calmodulin-like 3 |
| chr10_+_118305435 | 0.70 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr11_-_59612969 | 0.70 |
ENST00000541311.1
ENST00000257248.2 |
GIF
|
gastric intrinsic factor (vitamin B synthesis) |
| chr12_+_9102632 | 0.70 |
ENST00000539240.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr1_-_167487758 | 0.69 |
ENST00000362089.5
|
CD247
|
CD247 molecule |
| chr13_+_31309645 | 0.69 |
ENST00000380490.3
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein |
| chr20_-_54967187 | 0.69 |
ENST00000422322.1
ENST00000371356.2 ENST00000451915.1 ENST00000347343.2 ENST00000395911.1 ENST00000395907.1 ENST00000441357.1 ENST00000456249.1 ENST00000420474.1 ENST00000395909.4 ENST00000395914.1 ENST00000312783.6 ENST00000395915.3 ENST00000395913.3 |
AURKA
|
aurora kinase A |
| chr1_+_209859510 | 0.69 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr12_+_53662073 | 0.68 |
ENST00000553219.1
ENST00000257934.4 |
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr10_+_52152766 | 0.68 |
ENST00000596442.1
|
AC069547.2
|
Uncharacterized protein |
| chr19_-_51523275 | 0.67 |
ENST00000309958.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr12_-_10022735 | 0.66 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr4_-_52883786 | 0.66 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chrX_+_48380205 | 0.65 |
ENST00000446158.1
ENST00000414061.1 |
EBP
|
emopamil binding protein (sterol isomerase) |
| chr2_+_28618532 | 0.65 |
ENST00000545753.1
|
FOSL2
|
FOS-like antigen 2 |
| chr4_-_143227088 | 0.64 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr14_-_85996332 | 0.64 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr12_+_53662110 | 0.63 |
ENST00000552462.1
|
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr19_-_41934635 | 0.62 |
ENST00000321702.2
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr1_-_205912577 | 0.61 |
ENST00000367135.3
ENST00000367134.2 |
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr5_+_159848854 | 0.61 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr6_+_167704838 | 0.61 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr2_+_197504278 | 0.61 |
ENST00000272831.7
ENST00000389175.4 ENST00000472405.2 ENST00000423093.2 |
CCDC150
|
coiled-coil domain containing 150 |
| chr4_-_48082192 | 0.60 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr1_-_226187013 | 0.60 |
ENST00000272091.7
|
SDE2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr2_-_106054952 | 0.60 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr14_-_81893734 | 0.59 |
ENST00000555447.1
|
STON2
|
stonin 2 |
| chr7_+_155090271 | 0.59 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr17_-_64225508 | 0.59 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr20_+_34713312 | 0.58 |
ENST00000373946.3
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr12_+_75874984 | 0.58 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr2_+_210517895 | 0.57 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr3_+_101818088 | 0.57 |
ENST00000491959.1
|
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr19_-_36822595 | 0.57 |
ENST00000585356.1
ENST00000438368.2 ENST00000590622.1 |
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chr17_+_46537697 | 0.57 |
ENST00000579972.1
|
RP11-433M22.2
|
RP11-433M22.2 |
| chr1_+_115572415 | 0.56 |
ENST00000256592.1
|
TSHB
|
thyroid stimulating hormone, beta |
| chr10_-_71993176 | 0.56 |
ENST00000373232.3
|
PPA1
|
pyrophosphatase (inorganic) 1 |
| chr19_-_51017881 | 0.56 |
ENST00000601207.1
ENST00000598657.1 ENST00000376916.3 |
ASPDH
|
aspartate dehydrogenase domain containing |
| chr1_-_225616515 | 0.56 |
ENST00000338179.2
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr14_-_24584138 | 0.56 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr1_-_25747283 | 0.55 |
ENST00000346452.4
ENST00000340849.4 ENST00000349438.4 ENST00000294413.7 ENST00000413854.1 ENST00000455194.1 ENST00000243186.6 ENST00000425135.1 |
RHCE
|
Rh blood group, CcEe antigens |
| chr15_+_67418047 | 0.55 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr10_-_48416849 | 0.55 |
ENST00000249598.1
|
GDF2
|
growth differentiation factor 2 |
| chr1_+_192127578 | 0.55 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr15_+_42066632 | 0.55 |
ENST00000457542.2
ENST00000221214.6 ENST00000260357.7 ENST00000456763.2 |
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr6_+_31691121 | 0.55 |
ENST00000480039.1
ENST00000375810.4 ENST00000375805.2 ENST00000375809.3 ENST00000375804.2 ENST00000375814.3 ENST00000375806.2 |
C6orf25
|
chromosome 6 open reading frame 25 |
| chr2_-_231825668 | 0.54 |
ENST00000392039.2
|
GPR55
|
G protein-coupled receptor 55 |
| chr1_-_155270770 | 0.54 |
ENST00000392414.3
|
PKLR
|
pyruvate kinase, liver and RBC |
| chr1_-_159880159 | 0.53 |
ENST00000599780.1
|
AL590560.1
|
HCG1995379; Uncharacterized protein |
| chr11_+_128563652 | 0.53 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_+_167704798 | 0.53 |
ENST00000230256.3
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr1_+_35247859 | 0.53 |
ENST00000373362.3
|
GJB3
|
gap junction protein, beta 3, 31kDa |
| chr12_-_68647281 | 0.53 |
ENST00000328087.4
ENST00000538666.1 |
IL22
|
interleukin 22 |
| chr8_+_27629459 | 0.53 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr14_+_60975644 | 0.53 |
ENST00000327720.5
|
SIX6
|
SIX homeobox 6 |
| chr19_+_6739662 | 0.53 |
ENST00000313285.8
ENST00000313244.9 ENST00000596758.1 |
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr1_-_175162048 | 0.53 |
ENST00000444639.1
|
KIAA0040
|
KIAA0040 |
| chr13_+_32313658 | 0.52 |
ENST00000380314.1
ENST00000298386.2 |
RXFP2
|
relaxin/insulin-like family peptide receptor 2 |
| chr8_-_133123406 | 0.52 |
ENST00000434736.2
|
HHLA1
|
HERV-H LTR-associating 1 |
| chr1_+_207038699 | 0.52 |
ENST00000367098.1
|
IL20
|
interleukin 20 |
| chr8_-_15095832 | 0.52 |
ENST00000382080.1
|
SGCZ
|
sarcoglycan, zeta |
| chr1_-_167487808 | 0.51 |
ENST00000392122.3
|
CD247
|
CD247 molecule |
| chr7_+_123295861 | 0.51 |
ENST00000458573.2
ENST00000456238.2 |
LMOD2
|
leiomodin 2 (cardiac) |
| chr5_+_131409476 | 0.51 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr15_+_74165945 | 0.51 |
ENST00000535547.2
ENST00000300504.2 ENST00000562056.1 |
TBC1D21
|
TBC1 domain family, member 21 |
| chr6_-_112194484 | 0.51 |
ENST00000518295.1
ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr4_-_65275162 | 0.50 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr1_+_25599018 | 0.50 |
ENST00000417538.2
ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD
|
Rh blood group, D antigen |
| chr2_+_201994042 | 0.50 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr12_-_10324716 | 0.50 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chrX_-_55057403 | 0.50 |
ENST00000396198.3
ENST00000335854.4 ENST00000455688.1 ENST00000330807.5 |
ALAS2
|
aminolevulinate, delta-, synthase 2 |
| chr2_-_8464760 | 0.50 |
ENST00000430192.1
|
LINC00299
|
long intergenic non-protein coding RNA 299 |
| chr21_+_39628852 | 0.49 |
ENST00000398938.2
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr1_+_150954493 | 0.49 |
ENST00000368947.4
|
ANXA9
|
annexin A9 |
| chr17_-_57229155 | 0.49 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr17_+_37824217 | 0.49 |
ENST00000394246.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chrX_-_24665353 | 0.48 |
ENST00000379144.2
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr2_+_220436917 | 0.48 |
ENST00000243786.2
|
INHA
|
inhibin, alpha |
| chr14_-_72458326 | 0.48 |
ENST00000542853.1
|
AC005477.1
|
AC005477.1 |
| chr2_-_14541060 | 0.48 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr1_-_214638146 | 0.48 |
ENST00000543945.1
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr3_-_194188956 | 0.47 |
ENST00000256031.4
ENST00000446356.1 |
ATP13A3
|
ATPase type 13A3 |
| chr7_+_76139741 | 0.47 |
ENST00000334348.3
ENST00000419923.2 ENST00000448265.3 ENST00000443097.2 |
UPK3B
|
uroplakin 3B |
| chr22_-_29784519 | 0.47 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr10_-_25241499 | 0.46 |
ENST00000376378.1
ENST00000376376.3 ENST00000320152.6 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr2_+_90077680 | 0.46 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr5_-_76788317 | 0.46 |
ENST00000296679.4
|
WDR41
|
WD repeat domain 41 |
| chr2_+_234600253 | 0.46 |
ENST00000373424.1
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr17_+_4853442 | 0.45 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr6_-_107235331 | 0.45 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr11_-_19262486 | 0.45 |
ENST00000250024.4
|
E2F8
|
E2F transcription factor 8 |
| chr4_+_74606223 | 0.45 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr6_+_126661253 | 0.45 |
ENST00000368326.1
ENST00000368325.1 ENST00000368328.4 |
CENPW
|
centromere protein W |
| chr20_+_42984330 | 0.44 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr15_-_30113676 | 0.44 |
ENST00000400011.2
|
TJP1
|
tight junction protein 1 |
| chr1_+_50574585 | 0.44 |
ENST00000371824.1
ENST00000371823.4 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr6_-_35765088 | 0.44 |
ENST00000259938.2
|
CLPS
|
colipase, pancreatic |
| chr2_-_30144432 | 0.44 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr9_-_127269661 | 0.44 |
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr10_+_33271469 | 0.44 |
ENST00000414157.1
|
RP11-462L8.1
|
RP11-462L8.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA1_GATA4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.9 | GO:0002786 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.8 | 5.0 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.7 | 2.2 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.5 | 1.6 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.5 | 1.4 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.4 | 1.2 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.4 | 1.9 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.4 | 4.0 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.3 | 1.0 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.3 | 1.3 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.3 | 1.3 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.3 | 1.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.3 | 2.6 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.3 | 0.8 | GO:0060752 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 0.3 | 1.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.2 | 1.5 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 | 0.7 | GO:1900195 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 0.2 | 0.9 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 1.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 6.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 2.1 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.2 | 0.8 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.2 | 1.0 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.2 | 0.6 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 0.8 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.2 | 0.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 1.0 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 0.6 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.2 | 1.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 0.9 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.2 | 0.2 | GO:0002652 | regulation of tolerance induction dependent upon immune response(GO:0002652) |
| 0.2 | 0.7 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.2 | 0.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.2 | 0.5 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.2 | 0.6 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 3.9 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.4 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.4 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.4 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 | 0.6 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.1 | 0.6 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.7 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 0.5 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 0.4 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 1.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.6 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.9 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.3 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 1.3 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.9 | GO:0072205 | metanephric collecting duct development(GO:0072205) |
| 0.1 | 0.2 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.4 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.4 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 1.1 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 1.1 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.1 | 0.3 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.6 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.6 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 1.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 1.4 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.4 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.1 | 0.3 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.5 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 11.3 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.3 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.3 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 | 0.2 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.1 | 0.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.2 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.1 | 1.4 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.1 | 1.6 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.1 | 1.2 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.1 | 3.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.4 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.2 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.7 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 1.2 | GO:0048681 | neuronal signal transduction(GO:0023041) negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.8 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.2 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 0.4 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.1 | 0.5 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.4 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.3 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.1 | 1.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.3 | GO:2001027 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.3 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.7 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.2 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.1 | 0.5 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.2 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) renal water absorption(GO:0070295) |
| 0.1 | 0.9 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.1 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.2 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.3 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.3 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.3 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.8 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.9 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 1.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.2 | GO:1904346 | positive regulation of growth rate(GO:0040010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.1 | 0.5 | GO:0010629 | negative regulation of gene expression(GO:0010629) |
| 0.1 | 0.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 1.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 4.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.9 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 | 0.1 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.8 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.5 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 3.5 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.4 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.8 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 1.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.7 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 2.2 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.3 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.5 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.4 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.5 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.0 | 1.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 1.9 | GO:2000816 | negative regulation of sister chromatid segregation(GO:0033046) negative regulation of mitotic sister chromatid segregation(GO:0033048) negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.0 | 1.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.8 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.2 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.3 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.2 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.4 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.4 | GO:0070970 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) interleukin-2 secretion(GO:0070970) |
| 0.0 | 0.6 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 1.1 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) negative regulation of calcium-transporting ATPase activity(GO:1901895) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.0 | 0.2 | GO:0033384 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.3 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.7 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.4 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.5 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.8 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 2.1 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 1.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.4 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.1 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.0 | 0.6 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 1.8 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.8 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) |
| 0.0 | 0.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.0 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.5 | GO:0061621 | NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.5 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0019058 | viral life cycle(GO:0019058) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.4 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.1 | GO:0051873 | killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.5 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.9 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.2 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 1.0 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 3.5 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0070627 | ferrous iron import(GO:0070627) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.5 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.1 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 0.6 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.0 | GO:0035377 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.2 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.4 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.1 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.0 | 0.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.0 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.3 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 0.3 | GO:0001501 | skeletal system development(GO:0001501) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.2 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.8 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.5 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.5 | GO:0006953 | acute-phase response(GO:0006953) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.9 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.6 | 3.9 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.3 | 1.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 0.9 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.2 | 0.7 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.2 | 0.9 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 0.7 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.2 | 0.5 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 0.6 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 1.0 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 2.6 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.1 | 1.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 7.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.3 | GO:0097679 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) other organism cytoplasm(GO:0097679) |
| 0.1 | 0.9 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.3 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.3 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 1.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.3 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.1 | 1.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.4 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) 3M complex(GO:1990393) |
| 0.1 | 1.1 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 4.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.5 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.9 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 3.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 1.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 1.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.4 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.1 | GO:0036027 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 2.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 3.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.0 | 0.8 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 4.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 7.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.7 | 4.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.4 | 1.3 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.4 | 1.2 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.4 | 2.2 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.3 | 0.9 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.3 | 0.9 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.3 | 0.8 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.2 | 1.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 1.5 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.2 | 0.7 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.2 | 0.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 0.7 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.2 | 3.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 0.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 1.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 1.0 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 0.9 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 0.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 1.9 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 1.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 0.5 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.2 | 1.5 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.2 | 0.6 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 0.9 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.2 | 0.9 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 1.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.9 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.6 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 1.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.5 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 3.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.0 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 2.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.6 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.5 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.6 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.2 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 2.6 | GO:0005123 | death receptor binding(GO:0005123) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.2 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.1 | 0.7 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 1.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.4 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.5 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 2.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.2 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 0.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 1.1 | GO:0033612 | receptor serine/threonine kinase binding(GO:0033612) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.2 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 0.2 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.0 | 15.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.4 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.0 | 0.6 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 1.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 3.0 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 0.0 | 1.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 2.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.4 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 1.0 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 2.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 3.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 1.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 1.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.4 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 1.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 16.3 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 2.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.4 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.0 | GO:0015265 | urea transmembrane transporter activity(GO:0015204) urea channel activity(GO:0015265) amide transmembrane transporter activity(GO:0042887) |
| 0.0 | 0.0 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.5 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.2 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.8 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.5 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 3.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 5.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 1.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 2.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 3.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 6.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 7.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 3.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 3.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 3.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 4.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 2.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 2.5 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 2.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.8 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 2.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.8 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.4 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 2.1 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |