Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
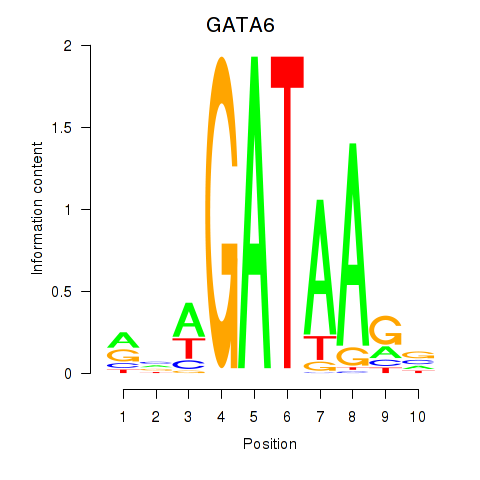
Results for GATA6
Z-value: 0.54
Transcription factors associated with GATA6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA6
|
ENSG00000141448.4 | GATA binding protein 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA6 | hg19_v2_chr18_+_19749386_19749404 | 0.57 | 1.0e-03 | Click! |
Activity profile of GATA6 motif
Sorted Z-values of GATA6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_102928009 | 9.26 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr9_+_33795533 | 1.77 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr17_+_74381343 | 1.51 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr20_-_54967187 | 1.37 |
ENST00000422322.1
ENST00000371356.2 ENST00000451915.1 ENST00000347343.2 ENST00000395911.1 ENST00000395907.1 ENST00000441357.1 ENST00000456249.1 ENST00000420474.1 ENST00000395909.4 ENST00000395914.1 ENST00000312783.6 ENST00000395915.3 ENST00000395913.3 |
AURKA
|
aurora kinase A |
| chrX_+_37865804 | 1.23 |
ENST00000297875.2
ENST00000357972.5 |
SYTL5
|
synaptotagmin-like 5 |
| chr6_+_12290586 | 1.05 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr18_+_19749386 | 1.02 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr1_-_94050668 | 0.97 |
ENST00000539242.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr7_+_142457315 | 0.93 |
ENST00000486171.1
ENST00000311737.7 |
PRSS1
|
protease, serine, 1 (trypsin 1) |
| chr12_+_4430371 | 0.83 |
ENST00000179259.4
|
C12orf5
|
chromosome 12 open reading frame 5 |
| chr4_-_103266355 | 0.80 |
ENST00000424970.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr7_+_155090271 | 0.78 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr5_+_131409476 | 0.77 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr1_-_24469602 | 0.73 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr5_+_159848807 | 0.69 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chrX_+_107288239 | 0.65 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chrX_+_107288197 | 0.65 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr2_-_69098566 | 0.63 |
ENST00000295379.1
|
BMP10
|
bone morphogenetic protein 10 |
| chr6_-_86099898 | 0.57 |
ENST00000455071.1
|
RP11-30P6.6
|
RP11-30P6.6 |
| chr9_-_114937465 | 0.56 |
ENST00000355396.3
|
SUSD1
|
sushi domain containing 1 |
| chr9_-_114937543 | 0.55 |
ENST00000374264.2
ENST00000374263.3 |
SUSD1
|
sushi domain containing 1 |
| chr9_-_114937676 | 0.49 |
ENST00000374270.3
|
SUSD1
|
sushi domain containing 1 |
| chr2_-_44065889 | 0.49 |
ENST00000543989.1
ENST00000405322.1 |
ABCG5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr2_-_44065946 | 0.47 |
ENST00000260645.1
|
ABCG5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr19_+_10397621 | 0.47 |
ENST00000380770.3
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr1_+_50574585 | 0.47 |
ENST00000371824.1
ENST00000371823.4 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr2_+_138722028 | 0.46 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr19_+_10397648 | 0.46 |
ENST00000340992.4
ENST00000393717.2 |
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr4_-_103266219 | 0.46 |
ENST00000394833.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr10_-_129691195 | 0.45 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
| chr13_-_46716969 | 0.44 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr1_-_158656488 | 0.43 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr3_-_47950745 | 0.41 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr11_-_66206260 | 0.39 |
ENST00000329819.4
ENST00000310999.7 ENST00000430466.2 |
MRPL11
|
mitochondrial ribosomal protein L11 |
| chr17_+_75283973 | 0.39 |
ENST00000431235.2
ENST00000449803.2 |
SEPT9
|
septin 9 |
| chr22_-_20231207 | 0.38 |
ENST00000425986.1
|
RTN4R
|
reticulon 4 receptor |
| chr11_+_10326612 | 0.34 |
ENST00000534464.1
ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM
|
adrenomedullin |
| chr6_+_130339710 | 0.34 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr17_+_19314505 | 0.33 |
ENST00000461366.1
|
RNF112
|
ring finger protein 112 |
| chr1_-_167487758 | 0.31 |
ENST00000362089.5
|
CD247
|
CD247 molecule |
| chr1_-_226187013 | 0.31 |
ENST00000272091.7
|
SDE2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr1_-_167487808 | 0.28 |
ENST00000392122.3
|
CD247
|
CD247 molecule |
| chr8_-_95449155 | 0.28 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr2_-_219850277 | 0.27 |
ENST00000295727.1
|
FEV
|
FEV (ETS oncogene family) |
| chr21_-_39870339 | 0.27 |
ENST00000429727.2
ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr8_+_21823726 | 0.26 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr11_+_118958689 | 0.26 |
ENST00000535253.1
ENST00000392841.1 |
HMBS
|
hydroxymethylbilane synthase |
| chr18_-_28742813 | 0.26 |
ENST00000257197.3
ENST00000257198.5 |
DSC1
|
desmocollin 1 |
| chr9_+_118950325 | 0.25 |
ENST00000534838.1
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr18_+_43304137 | 0.23 |
ENST00000502059.2
ENST00000586951.1 ENST00000589322.2 ENST00000415427.3 ENST00000535474.1 ENST00000402943.2 |
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr19_-_13213662 | 0.23 |
ENST00000264824.4
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr12_-_52995291 | 0.23 |
ENST00000293745.2
ENST00000354310.4 |
KRT72
|
keratin 72 |
| chr2_+_102927962 | 0.23 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr1_+_66999799 | 0.22 |
ENST00000371035.3
ENST00000371036.3 ENST00000371037.4 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr7_-_100239132 | 0.20 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr10_+_71075552 | 0.19 |
ENST00000298649.3
|
HK1
|
hexokinase 1 |
| chr1_-_42630389 | 0.19 |
ENST00000357001.2
|
GUCA2A
|
guanylate cyclase activator 2A (guanylin) |
| chr2_+_87565634 | 0.18 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr3_+_148583043 | 0.18 |
ENST00000296046.3
|
CPA3
|
carboxypeptidase A3 (mast cell) |
| chr2_-_90538397 | 0.16 |
ENST00000443397.3
|
RP11-685N3.1
|
Uncharacterized protein |
| chr12_-_120765565 | 0.15 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr1_-_120354079 | 0.15 |
ENST00000354219.1
ENST00000369401.4 ENST00000256585.5 |
REG4
|
regenerating islet-derived family, member 4 |
| chr1_+_25599018 | 0.14 |
ENST00000417538.2
ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD
|
Rh blood group, D antigen |
| chr8_+_70404996 | 0.13 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr10_+_118350468 | 0.13 |
ENST00000358834.4
ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr1_+_206317450 | 0.13 |
ENST00000358184.2
ENST00000361052.3 ENST00000360218.2 |
CTSE
|
cathepsin E |
| chr19_-_11494975 | 0.12 |
ENST00000222139.6
ENST00000592375.2 |
EPOR
|
erythropoietin receptor |
| chr22_-_29784519 | 0.12 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr1_+_12524965 | 0.12 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr4_+_15376165 | 0.12 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr12_+_70219052 | 0.11 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr19_-_51522955 | 0.09 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr5_-_135290705 | 0.08 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr10_+_118350522 | 0.08 |
ENST00000530319.1
ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr2_-_152830441 | 0.08 |
ENST00000534999.1
ENST00000397327.2 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr6_-_30128657 | 0.07 |
ENST00000449742.2
ENST00000376704.3 |
TRIM10
|
tripartite motif containing 10 |
| chr5_+_1201703 | 0.07 |
ENST00000304460.10
|
SLC6A19
|
solute carrier family 6 (neutral amino acid transporter), member 19 |
| chr17_+_4835580 | 0.06 |
ENST00000329125.5
|
GP1BA
|
glycoprotein Ib (platelet), alpha polypeptide |
| chr20_-_30795511 | 0.06 |
ENST00000246229.4
|
PLAGL2
|
pleiomorphic adenoma gene-like 2 |
| chr5_-_33984741 | 0.06 |
ENST00000382102.3
ENST00000509381.1 ENST00000342059.3 ENST00000345083.5 |
SLC45A2
|
solute carrier family 45, member 2 |
| chr5_+_135496675 | 0.06 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr4_+_100495864 | 0.06 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr1_+_25598872 | 0.05 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
| chr1_+_25598989 | 0.05 |
ENST00000454452.2
|
RHD
|
Rh blood group, D antigen |
| chr5_-_33984786 | 0.04 |
ENST00000296589.4
|
SLC45A2
|
solute carrier family 45, member 2 |
| chr3_+_137717571 | 0.04 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chr18_+_43304092 | 0.04 |
ENST00000321925.4
ENST00000587601.1 |
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr17_-_4806369 | 0.03 |
ENST00000293780.4
|
CHRNE
|
cholinergic receptor, nicotinic, epsilon (muscle) |
| chr11_-_26743546 | 0.03 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr12_-_6233828 | 0.03 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr6_+_31674639 | 0.03 |
ENST00000556581.1
ENST00000375832.4 ENST00000503322.1 |
LY6G6F
MEGT1
|
lymphocyte antigen 6 complex, locus G6F HCG43720, isoform CRA_a; Lymphocyte antigen 6 complex locus protein G6f; Megakaryocyte-enhanced gene transcript 1 protein; Uncharacterized protein |
| chr20_+_54967409 | 0.03 |
ENST00000415828.1
ENST00000217109.4 ENST00000428552.1 |
CSTF1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa |
| chr8_+_70378852 | 0.03 |
ENST00000525061.1
ENST00000458141.2 ENST00000260128.4 |
SULF1
|
sulfatase 1 |
| chr15_+_49715293 | 0.03 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr19_+_14138960 | 0.02 |
ENST00000431365.2
ENST00000585987.1 |
RLN3
|
relaxin 3 |
| chr12_-_56221330 | 0.01 |
ENST00000546837.1
|
RP11-762I7.5
|
Uncharacterized protein |
| chr15_+_59279851 | 0.01 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr12_+_72332618 | 0.01 |
ENST00000333850.3
|
TPH2
|
tryptophan hydroxylase 2 |
| chr17_-_38938786 | 0.01 |
ENST00000301656.3
|
KRT27
|
keratin 27 |
| chr14_+_21945335 | 0.01 |
ENST00000262709.3
ENST00000457430.2 ENST00000448790.2 |
TOX4
|
TOX high mobility group box family member 4 |
| chr7_+_128379346 | 0.01 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr2_+_220436917 | 0.00 |
ENST00000243786.2
|
INHA
|
inhibin, alpha |
| chr3_+_69812877 | 0.00 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr17_-_62084241 | 0.00 |
ENST00000449662.2
|
ICAM2
|
intercellular adhesion molecule 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 9.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.5 | 1.5 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.5 | 1.4 | GO:1900195 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 0.3 | 1.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.3 | 1.0 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.3 | 0.8 | GO:0045796 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 0.3 | 0.8 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 0.8 | GO:0045918 | positive regulation of interleukin-23 production(GO:0032747) negative regulation of cytolysis(GO:0045918) |
| 0.1 | 1.3 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.6 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.1 | 2.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.5 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.3 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.3 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.2 | GO:0070627 | ferrous iron import(GO:0070627) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 1.0 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.4 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 1.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.3 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.0 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.7 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.0 | 0.2 | GO:0002003 | angiotensin maturation(GO:0002003) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.3 | 1.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 0.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.8 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 10.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.3 | 1.5 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 1.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 1.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.4 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.3 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 0.2 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 1.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 3.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.0 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |