Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
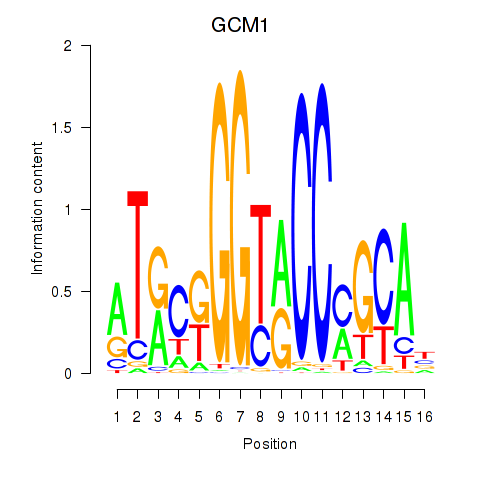
Results for GCM1
Z-value: 0.65
Transcription factors associated with GCM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GCM1
|
ENSG00000137270.10 | glial cells missing transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GCM1 | hg19_v2_chr6_-_53013620_53013644 | -0.21 | 2.7e-01 | Click! |
Activity profile of GCM1 motif
Sorted Z-values of GCM1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_57547454 | 1.34 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr4_-_57547870 | 1.33 |
ENST00000381260.3
ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX
|
HOP homeobox |
| chr2_+_90108504 | 1.25 |
ENST00000390271.2
|
IGKV6D-41
|
immunoglobulin kappa variable 6D-41 (non-functional) |
| chr11_+_13690249 | 1.05 |
ENST00000532701.1
|
FAR1
|
fatty acyl CoA reductase 1 |
| chr2_+_90060377 | 1.04 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr2_-_89459813 | 0.98 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr20_-_1306351 | 0.98 |
ENST00000381812.1
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr20_+_361261 | 0.97 |
ENST00000217233.3
|
TRIB3
|
tribbles pseudokinase 3 |
| chr20_-_1306391 | 0.95 |
ENST00000339987.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr12_-_122238464 | 0.88 |
ENST00000546227.1
|
RHOF
|
ras homolog family member F (in filopodia) |
| chr17_-_7493390 | 0.84 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr9_+_131182697 | 0.82 |
ENST00000372838.4
ENST00000411852.1 |
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr3_+_10206545 | 0.81 |
ENST00000256458.4
|
IRAK2
|
interleukin-1 receptor-associated kinase 2 |
| chr14_+_24540046 | 0.72 |
ENST00000397016.2
ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6
|
copine VI (neuronal) |
| chr22_+_22936998 | 0.69 |
ENST00000390303.2
|
IGLV3-32
|
immunoglobulin lambda variable 3-32 (non-functional) |
| chr18_+_21529811 | 0.67 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr22_-_20256054 | 0.66 |
ENST00000043402.7
|
RTN4R
|
reticulon 4 receptor |
| chr11_+_111126707 | 0.65 |
ENST00000280325.4
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr11_-_8190534 | 0.62 |
ENST00000309737.6
ENST00000425599.2 ENST00000539720.1 ENST00000531450.1 ENST00000419822.2 ENST00000335425.7 ENST00000343202.4 |
RIC3
|
RIC3 acetylcholine receptor chaperone |
| chr14_-_24732368 | 0.62 |
ENST00000544573.1
|
TGM1
|
transglutaminase 1 |
| chr22_+_22786288 | 0.59 |
ENST00000390301.2
|
IGLV1-36
|
immunoglobulin lambda variable 1-36 |
| chr11_+_844067 | 0.59 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr4_-_56502426 | 0.58 |
ENST00000505262.1
ENST00000507338.1 |
NMU
|
neuromedin U |
| chr18_+_21269556 | 0.50 |
ENST00000399516.3
|
LAMA3
|
laminin, alpha 3 |
| chr17_-_14140166 | 0.48 |
ENST00000420162.2
ENST00000431716.2 |
CDRT15
|
CMT1A duplicated region transcript 15 |
| chr22_-_20231207 | 0.48 |
ENST00000425986.1
|
RTN4R
|
reticulon 4 receptor |
| chr16_-_755726 | 0.47 |
ENST00000324361.5
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr22_-_20255212 | 0.46 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr11_+_13690200 | 0.45 |
ENST00000354817.3
|
FAR1
|
fatty acyl CoA reductase 1 |
| chr4_-_56502451 | 0.45 |
ENST00000511469.1
ENST00000264218.3 |
NMU
|
neuromedin U |
| chr19_-_48059113 | 0.44 |
ENST00000391901.3
ENST00000314121.4 ENST00000448976.1 |
ZNF541
|
zinc finger protein 541 |
| chr12_-_69326940 | 0.43 |
ENST00000549781.1
ENST00000548262.1 ENST00000551568.1 ENST00000548954.1 |
CPM
|
carboxypeptidase M |
| chr22_+_44319648 | 0.42 |
ENST00000423180.2
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr18_+_21269404 | 0.42 |
ENST00000313654.9
|
LAMA3
|
laminin, alpha 3 |
| chr14_+_24540731 | 0.41 |
ENST00000558859.1
ENST00000559197.1 ENST00000560828.1 ENST00000216775.2 ENST00000560884.1 |
CPNE6
|
copine VI (neuronal) |
| chr7_-_100253993 | 0.41 |
ENST00000461605.1
ENST00000160382.5 |
ACTL6B
|
actin-like 6B |
| chr17_+_40913264 | 0.40 |
ENST00000587142.1
ENST00000588576.1 |
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr9_+_4985016 | 0.39 |
ENST00000539801.1
|
JAK2
|
Janus kinase 2 |
| chr10_-_43762329 | 0.39 |
ENST00000395810.1
|
RASGEF1A
|
RasGEF domain family, member 1A |
| chr10_+_15074190 | 0.37 |
ENST00000428897.1
ENST00000413672.1 |
OLAH
|
oleoyl-ACP hydrolase |
| chr16_-_755819 | 0.35 |
ENST00000397621.1
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr1_-_40237020 | 0.35 |
ENST00000327582.5
|
OXCT2
|
3-oxoacid CoA transferase 2 |
| chr11_+_35198118 | 0.35 |
ENST00000525211.1
ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr18_-_5419797 | 0.35 |
ENST00000542146.1
ENST00000427684.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr11_+_124735282 | 0.34 |
ENST00000397801.1
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr4_-_8073705 | 0.34 |
ENST00000514025.1
|
ABLIM2
|
actin binding LIM protein family, member 2 |
| chr17_+_80517216 | 0.33 |
ENST00000531030.1
ENST00000526383.2 |
FOXK2
|
forkhead box K2 |
| chr16_-_74734672 | 0.32 |
ENST00000306247.7
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr2_+_47168630 | 0.32 |
ENST00000263737.6
|
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr3_+_12329397 | 0.32 |
ENST00000397015.2
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr19_-_3480540 | 0.32 |
ENST00000215531.4
|
C19orf77
|
chromosome 19 open reading frame 77 |
| chr10_-_73975657 | 0.31 |
ENST00000394919.1
ENST00000526751.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr4_-_987217 | 0.31 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr3_+_12329358 | 0.31 |
ENST00000309576.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr19_-_52227221 | 0.30 |
ENST00000222115.1
ENST00000540069.2 |
HAS1
|
hyaluronan synthase 1 |
| chr1_+_25071848 | 0.30 |
ENST00000374379.4
|
CLIC4
|
chloride intracellular channel 4 |
| chr6_+_30029008 | 0.29 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr10_-_73976025 | 0.29 |
ENST00000342444.4
ENST00000533958.1 ENST00000527593.1 ENST00000394915.3 ENST00000530461.1 ENST00000317168.6 ENST00000524829.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr4_-_40859132 | 0.28 |
ENST00000543538.1
ENST00000502841.1 ENST00000504305.1 ENST00000513516.1 ENST00000510670.1 |
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr16_-_74734742 | 0.28 |
ENST00000308807.7
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr12_+_8666126 | 0.28 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr19_+_12902289 | 0.28 |
ENST00000302754.4
|
JUNB
|
jun B proto-oncogene |
| chr18_+_10526008 | 0.28 |
ENST00000542979.1
ENST00000322897.6 |
NAPG
|
N-ethylmaleimide-sensitive factor attachment protein, gamma |
| chr10_-_105677427 | 0.27 |
ENST00000369764.1
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr8_-_144099795 | 0.27 |
ENST00000522060.1
ENST00000517833.1 ENST00000502167.2 ENST00000518831.1 |
RP11-273G15.2
|
RP11-273G15.2 |
| chr7_-_100860851 | 0.26 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr16_-_90096309 | 0.25 |
ENST00000408886.2
|
C16orf3
|
chromosome 16 open reading frame 3 |
| chr11_-_62783276 | 0.25 |
ENST00000535878.1
ENST00000545207.1 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr5_+_135385202 | 0.25 |
ENST00000514554.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr4_-_52904425 | 0.25 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr16_-_19729502 | 0.24 |
ENST00000219837.7
|
KNOP1
|
lysine-rich nucleolar protein 1 |
| chr9_+_17579084 | 0.24 |
ENST00000380607.4
|
SH3GL2
|
SH3-domain GRB2-like 2 |
| chr9_-_111619239 | 0.24 |
ENST00000374667.3
|
ACTL7B
|
actin-like 7B |
| chr1_+_44399466 | 0.24 |
ENST00000498139.2
ENST00000491846.1 |
ARTN
|
artemin |
| chr6_+_33589161 | 0.24 |
ENST00000605930.1
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr2_-_89630186 | 0.24 |
ENST00000390264.2
|
IGKV2-40
|
immunoglobulin kappa variable 2-40 |
| chr16_-_57513657 | 0.23 |
ENST00000566936.1
ENST00000568617.1 ENST00000567276.1 ENST00000569548.1 ENST00000569250.1 ENST00000564378.1 |
DOK4
|
docking protein 4 |
| chr19_+_11546440 | 0.23 |
ENST00000589126.1
ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr22_+_38864041 | 0.23 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr12_-_122238913 | 0.22 |
ENST00000537157.1
|
AC084018.1
|
AC084018.1 |
| chr17_+_40913210 | 0.22 |
ENST00000253796.5
|
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chrX_-_152486108 | 0.22 |
ENST00000356661.5
|
MAGEA1
|
melanoma antigen family A, 1 (directs expression of antigen MZ2-E) |
| chr14_-_50778872 | 0.22 |
ENST00000555610.1
ENST00000261699.4 |
L2HGDH
|
L-2-hydroxyglutarate dehydrogenase |
| chr19_-_6379069 | 0.22 |
ENST00000597721.1
|
PSPN
|
persephin |
| chr20_+_33563206 | 0.21 |
ENST00000262873.7
|
MYH7B
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr19_-_38397228 | 0.21 |
ENST00000447313.2
|
WDR87
|
WD repeat domain 87 |
| chr7_-_26904317 | 0.20 |
ENST00000345317.2
|
SKAP2
|
src kinase associated phosphoprotein 2 |
| chr3_-_196242233 | 0.20 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr3_-_121468513 | 0.20 |
ENST00000494517.1
ENST00000393667.3 |
GOLGB1
|
golgin B1 |
| chr20_-_45061695 | 0.20 |
ENST00000445496.2
|
ELMO2
|
engulfment and cell motility 2 |
| chr3_-_121468602 | 0.20 |
ENST00000340645.5
|
GOLGB1
|
golgin B1 |
| chr8_+_25316707 | 0.20 |
ENST00000380665.3
|
CDCA2
|
cell division cycle associated 2 |
| chr5_-_133702761 | 0.20 |
ENST00000521118.1
ENST00000265334.4 ENST00000435211.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr8_+_142138799 | 0.20 |
ENST00000518668.1
|
DENND3
|
DENN/MADD domain containing 3 |
| chr16_+_56598961 | 0.20 |
ENST00000219162.3
|
MT4
|
metallothionein 4 |
| chr2_+_233320827 | 0.20 |
ENST00000295463.3
|
ALPI
|
alkaline phosphatase, intestinal |
| chr7_-_44179972 | 0.20 |
ENST00000446581.1
|
MYL7
|
myosin, light chain 7, regulatory |
| chr16_-_85722530 | 0.19 |
ENST00000253462.3
|
GINS2
|
GINS complex subunit 2 (Psf2 homolog) |
| chr1_-_11907829 | 0.19 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr17_-_40913275 | 0.19 |
ENST00000589716.1
ENST00000360166.3 |
RAMP2-AS1
|
RAMP2 antisense RNA 1 |
| chr19_-_38397285 | 0.18 |
ENST00000303868.5
|
WDR87
|
WD repeat domain 87 |
| chrX_-_27417088 | 0.18 |
ENST00000608735.1
ENST00000422048.1 |
RP11-268G12.1
|
RP11-268G12.1 |
| chr8_+_104383728 | 0.18 |
ENST00000330295.5
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr15_+_78632666 | 0.18 |
ENST00000299529.6
|
CRABP1
|
cellular retinoic acid binding protein 1 |
| chr19_+_11658655 | 0.17 |
ENST00000588935.1
|
CNN1
|
calponin 1, basic, smooth muscle |
| chr11_+_31531291 | 0.17 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr3_+_130613226 | 0.17 |
ENST00000509662.1
ENST00000328560.8 ENST00000428331.2 ENST00000359644.3 ENST00000422190.2 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr4_+_110749143 | 0.17 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr2_+_88469835 | 0.17 |
ENST00000358591.2
ENST00000377254.3 ENST00000402102.1 ENST00000419759.1 ENST00000449349.1 ENST00000343544.4 |
THNSL2
|
threonine synthase-like 2 (S. cerevisiae) |
| chr6_-_30043539 | 0.17 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr17_-_39140549 | 0.17 |
ENST00000377755.4
|
KRT40
|
keratin 40 |
| chr10_+_96162242 | 0.17 |
ENST00000225235.4
|
TBC1D12
|
TBC1 domain family, member 12 |
| chr9_-_115653176 | 0.17 |
ENST00000374228.4
|
SLC46A2
|
solute carrier family 46, member 2 |
| chr2_+_208527094 | 0.16 |
ENST00000429730.1
|
AC079767.4
|
AC079767.4 |
| chr13_-_78492927 | 0.16 |
ENST00000334286.5
|
EDNRB
|
endothelin receptor type B |
| chr10_-_105677886 | 0.16 |
ENST00000224950.3
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr3_-_128369643 | 0.16 |
ENST00000296255.3
|
RPN1
|
ribophorin I |
| chr19_+_641178 | 0.16 |
ENST00000166133.3
|
FGF22
|
fibroblast growth factor 22 |
| chr20_+_30225682 | 0.16 |
ENST00000376075.3
|
COX4I2
|
cytochrome c oxidase subunit IV isoform 2 (lung) |
| chr16_+_2285817 | 0.16 |
ENST00000564065.1
|
DNASE1L2
|
deoxyribonuclease I-like 2 |
| chr6_-_74363803 | 0.16 |
ENST00000355773.5
|
SLC17A5
|
solute carrier family 17 (acidic sugar transporter), member 5 |
| chr2_-_106810742 | 0.16 |
ENST00000409501.3
ENST00000428048.2 ENST00000441952.1 ENST00000457835.1 ENST00000540130.1 |
UXS1
|
UDP-glucuronate decarboxylase 1 |
| chr17_-_4269920 | 0.16 |
ENST00000572484.1
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr16_+_2022036 | 0.15 |
ENST00000568546.1
|
TBL3
|
transducin (beta)-like 3 |
| chr8_-_57232656 | 0.15 |
ENST00000396721.2
|
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5 |
| chr2_-_106810783 | 0.15 |
ENST00000283148.7
|
UXS1
|
UDP-glucuronate decarboxylase 1 |
| chr19_+_54705025 | 0.14 |
ENST00000441429.1
|
RPS9
|
ribosomal protein S9 |
| chr20_+_44441304 | 0.14 |
ENST00000352551.5
|
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr15_-_94443820 | 0.14 |
ENST00000557481.2
|
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr7_-_150780609 | 0.14 |
ENST00000297533.4
|
TMUB1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr12_-_39300071 | 0.14 |
ENST00000550863.1
|
CPNE8
|
copine VIII |
| chr6_-_136847610 | 0.14 |
ENST00000454590.1
ENST00000432797.2 |
MAP7
|
microtubule-associated protein 7 |
| chrX_-_84634737 | 0.14 |
ENST00000262753.4
|
POF1B
|
premature ovarian failure, 1B |
| chr1_-_2145620 | 0.14 |
ENST00000545087.1
|
AL590822.1
|
Uncharacterized protein |
| chr1_+_11796126 | 0.14 |
ENST00000376637.3
|
AGTRAP
|
angiotensin II receptor-associated protein |
| chr2_-_24583168 | 0.14 |
ENST00000361999.3
|
ITSN2
|
intersectin 2 |
| chr15_-_45422056 | 0.14 |
ENST00000267803.4
ENST00000559014.1 ENST00000558851.1 ENST00000559988.1 ENST00000558996.1 ENST00000558422.1 ENST00000559226.1 ENST00000558326.1 ENST00000558377.1 ENST00000559644.1 |
DUOXA1
|
dual oxidase maturation factor 1 |
| chr1_+_24069642 | 0.13 |
ENST00000418390.2
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A) |
| chr1_+_24069952 | 0.13 |
ENST00000609199.1
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A) |
| chr16_+_67062996 | 0.13 |
ENST00000561924.2
|
CBFB
|
core-binding factor, beta subunit |
| chr17_-_62340581 | 0.13 |
ENST00000258991.3
ENST00000583738.1 ENST00000584379.1 |
TEX2
|
testis expressed 2 |
| chr19_-_7797045 | 0.13 |
ENST00000328853.5
|
CLEC4G
|
C-type lectin domain family 4, member G |
| chrX_-_84634708 | 0.13 |
ENST00000373145.3
|
POF1B
|
premature ovarian failure, 1B |
| chr1_-_26394114 | 0.13 |
ENST00000374272.3
|
TRIM63
|
tripartite motif containing 63, E3 ubiquitin protein ligase |
| chr5_+_140767452 | 0.13 |
ENST00000519479.1
|
PCDHGB4
|
protocadherin gamma subfamily B, 4 |
| chr5_-_127674883 | 0.12 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr2_-_89385283 | 0.12 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr7_-_4901625 | 0.12 |
ENST00000404991.1
|
PAPOLB
|
poly(A) polymerase beta (testis specific) |
| chr9_+_138628365 | 0.12 |
ENST00000491806.2
ENST00000488444.2 ENST00000490355.2 ENST00000263604.3 |
KCNT1
|
potassium channel, subfamily T, member 1 |
| chr19_-_1650666 | 0.11 |
ENST00000588136.1
|
TCF3
|
transcription factor 3 |
| chr11_+_64058758 | 0.11 |
ENST00000538767.1
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr6_+_3068606 | 0.11 |
ENST00000259808.4
|
RIPK1
|
receptor (TNFRSF)-interacting serine-threonine kinase 1 |
| chr18_-_18691739 | 0.11 |
ENST00000399799.2
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr22_+_22930626 | 0.11 |
ENST00000390302.2
|
IGLV2-33
|
immunoglobulin lambda variable 2-33 (non-functional) |
| chr12_-_4554780 | 0.11 |
ENST00000228837.2
|
FGF6
|
fibroblast growth factor 6 |
| chr4_+_56212505 | 0.10 |
ENST00000505210.1
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr2_-_113993020 | 0.10 |
ENST00000465084.1
|
PAX8
|
paired box 8 |
| chr16_+_67063262 | 0.10 |
ENST00000565389.1
|
CBFB
|
core-binding factor, beta subunit |
| chr7_+_99971068 | 0.10 |
ENST00000198536.2
ENST00000453419.1 |
PILRA
|
paired immunoglobin-like type 2 receptor alpha |
| chr4_-_48782259 | 0.10 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr19_+_35842445 | 0.10 |
ENST00000246553.2
|
FFAR1
|
free fatty acid receptor 1 |
| chr20_+_36946029 | 0.10 |
ENST00000417318.1
|
BPI
|
bactericidal/permeability-increasing protein |
| chr14_-_54908043 | 0.10 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr1_+_11796177 | 0.10 |
ENST00000400895.2
ENST00000376629.4 ENST00000376627.2 ENST00000314340.5 ENST00000452018.2 ENST00000510878.1 |
AGTRAP
|
angiotensin II receptor-associated protein |
| chr17_-_4269768 | 0.09 |
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr9_-_99540328 | 0.09 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr4_+_56212270 | 0.09 |
ENST00000264228.4
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr19_+_54704718 | 0.09 |
ENST00000391752.1
ENST00000402367.1 ENST00000391751.3 |
RPS9
|
ribosomal protein S9 |
| chr17_-_3599492 | 0.09 |
ENST00000435558.1
ENST00000345901.3 |
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr4_-_119274121 | 0.09 |
ENST00000296498.3
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr15_-_78933567 | 0.09 |
ENST00000261751.3
ENST00000412074.2 |
CHRNB4
|
cholinergic receptor, nicotinic, beta 4 (neuronal) |
| chr11_+_20385231 | 0.09 |
ENST00000530266.1
ENST00000421577.2 ENST00000443524.2 ENST00000419348.2 |
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa |
| chr11_+_64058820 | 0.09 |
ENST00000422670.2
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr11_+_20385666 | 0.09 |
ENST00000532081.1
ENST00000531058.1 |
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa |
| chr2_+_234959323 | 0.08 |
ENST00000373368.1
ENST00000168148.3 |
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr17_-_3599327 | 0.08 |
ENST00000551178.1
ENST00000552276.1 ENST00000547178.1 |
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr22_+_22599189 | 0.08 |
ENST00000302273.2
|
VPREB1
|
pre-B lymphocyte 1 |
| chr21_-_38445443 | 0.08 |
ENST00000360525.4
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr9_-_138591341 | 0.08 |
ENST00000298466.5
ENST00000425225.1 |
SOHLH1
|
spermatogenesis and oogenesis specific basic helix-loop-helix 1 |
| chr4_+_38511367 | 0.08 |
ENST00000507056.1
|
RP11-213G21.1
|
RP11-213G21.1 |
| chr19_-_1021113 | 0.08 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr19_+_50936142 | 0.08 |
ENST00000357701.5
|
MYBPC2
|
myosin binding protein C, fast type |
| chr7_-_50132860 | 0.08 |
ENST00000046087.2
|
ZPBP
|
zona pellucida binding protein |
| chr22_+_22764088 | 0.08 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr7_-_50132801 | 0.08 |
ENST00000419417.1
|
ZPBP
|
zona pellucida binding protein |
| chr19_+_54704990 | 0.08 |
ENST00000391753.2
|
RPS9
|
ribosomal protein S9 |
| chr1_-_1690014 | 0.08 |
ENST00000400922.2
ENST00000342348.5 |
NADK
|
NAD kinase |
| chr20_+_34043085 | 0.08 |
ENST00000397527.1
ENST00000342580.4 |
CEP250
|
centrosomal protein 250kDa |
| chr6_-_132032206 | 0.08 |
ENST00000314099.8
|
CTAGE9
|
CTAGE family, member 9 |
| chr3_+_46448648 | 0.08 |
ENST00000399036.3
|
CCRL2
|
chemokine (C-C motif) receptor-like 2 |
| chr3_+_130613001 | 0.07 |
ENST00000504948.1
ENST00000513801.1 ENST00000505072.1 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr20_+_30639991 | 0.07 |
ENST00000534862.1
ENST00000538448.1 ENST00000375862.2 |
HCK
|
hemopoietic cell kinase |
| chr7_+_100860949 | 0.07 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr8_-_97247759 | 0.07 |
ENST00000518406.1
ENST00000523920.1 ENST00000287022.5 |
UQCRB
|
ubiquinol-cytochrome c reductase binding protein |
| chr4_-_17783135 | 0.07 |
ENST00000265018.3
|
FAM184B
|
family with sequence similarity 184, member B |
| chr1_-_8938736 | 0.07 |
ENST00000234590.4
|
ENO1
|
enolase 1, (alpha) |
| chr1_-_159880159 | 0.07 |
ENST00000599780.1
|
AL590560.1
|
HCG1995379; Uncharacterized protein |
| chr7_+_143880559 | 0.07 |
ENST00000486333.1
|
CTAGE4
|
CTAGE family, member 4 |
| chr14_+_39736299 | 0.07 |
ENST00000341502.5
ENST00000396158.2 ENST00000280083.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr22_+_19705928 | 0.07 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GCM1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.3 | 0.8 | GO:0048627 | myoblast development(GO:0048627) |
| 0.2 | 0.6 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.2 | 0.6 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 3.0 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 1.0 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.3 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 1.6 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.3 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.4 | GO:1902728 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 0.6 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.2 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.1 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 1.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0007497 | posterior midgut development(GO:0007497) |
| 0.0 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.2 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 1.0 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.3 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.2 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.3 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) positive regulation of necroptotic process(GO:0060545) |
| 0.0 | 0.4 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.1 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.5 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 1.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.8 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.3 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 4.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:2001171 | positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.0 | 0.1 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.6 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.2 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.0 | 0.2 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.3 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.6 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 1.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 1.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 1.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.3 | 1.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.4 | GO:0016296 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.6 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.2 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.3 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.4 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.0 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 1.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.3 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 1.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.0 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.0 | GO:0070546 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 3.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 0.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 1.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |