Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
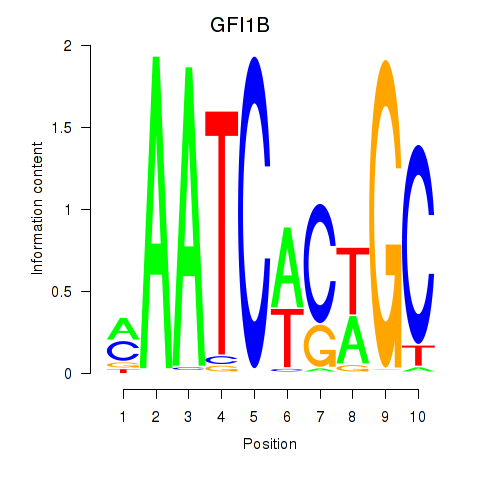
Results for GFI1B
Z-value: 0.56
Transcription factors associated with GFI1B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GFI1B
|
ENSG00000165702.8 | growth factor independent 1B transcriptional repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GFI1B | hg19_v2_chr9_+_135854091_135854159 | -0.29 | 1.2e-01 | Click! |
Activity profile of GFI1B motif
Sorted Z-values of GFI1B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_43916296 | 2.51 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr21_-_43916433 | 1.78 |
ENST00000291536.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr15_+_71228826 | 1.70 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr20_+_56725952 | 1.67 |
ENST00000371168.3
|
C20orf85
|
chromosome 20 open reading frame 85 |
| chr19_+_41620335 | 1.60 |
ENST00000331105.2
|
CYP2F1
|
cytochrome P450, family 2, subfamily F, polypeptide 1 |
| chr5_-_137475071 | 1.36 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr11_+_111385497 | 1.18 |
ENST00000375618.4
ENST00000529167.1 ENST00000332814.6 |
C11orf88
|
chromosome 11 open reading frame 88 |
| chr17_-_19281203 | 1.15 |
ENST00000487415.2
|
B9D1
|
B9 protein domain 1 |
| chr18_+_77905894 | 1.13 |
ENST00000589574.1
ENST00000588226.1 ENST00000585422.1 |
AC139100.2
|
Uncharacterized protein |
| chr4_+_15471489 | 1.10 |
ENST00000424120.1
ENST00000413206.1 ENST00000438599.2 ENST00000511544.1 ENST00000512702.1 ENST00000507954.1 ENST00000515124.1 ENST00000503292.1 ENST00000503658.1 |
CC2D2A
|
coiled-coil and C2 domain containing 2A |
| chr11_+_71791849 | 1.02 |
ENST00000423494.2
ENST00000539587.1 ENST00000538478.1 ENST00000324866.7 ENST00000439209.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr3_-_158450231 | 1.00 |
ENST00000479756.1
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr3_-_158450475 | 0.92 |
ENST00000237696.5
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr11_+_71791693 | 0.88 |
ENST00000289488.2
ENST00000447974.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr11_+_101785727 | 0.86 |
ENST00000263468.8
|
KIAA1377
|
KIAA1377 |
| chr5_+_36608422 | 0.84 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr5_+_140261703 | 0.82 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr11_+_71791803 | 0.79 |
ENST00000539271.1
|
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr4_+_39184024 | 0.76 |
ENST00000399820.3
ENST00000509560.1 ENST00000512112.1 ENST00000288634.7 ENST00000506503.1 |
WDR19
|
WD repeat domain 19 |
| chr19_-_51412584 | 0.75 |
ENST00000431178.2
|
KLK4
|
kallikrein-related peptidase 4 |
| chr7_-_6007070 | 0.71 |
ENST00000337579.3
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr7_+_6796986 | 0.71 |
ENST00000297186.3
|
RSPH10B2
|
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chr16_-_18441131 | 0.70 |
ENST00000339303.5
|
NPIPA8
|
nuclear pore complex interacting protein family, member A8 |
| chr7_-_150721570 | 0.70 |
ENST00000377974.2
ENST00000444312.1 ENST00000605938.1 ENST00000605952.1 |
ATG9B
|
autophagy related 9B |
| chr5_-_124080203 | 0.69 |
ENST00000504926.1
|
ZNF608
|
zinc finger protein 608 |
| chr11_+_27076764 | 0.68 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr5_-_96478466 | 0.63 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr4_+_41362796 | 0.60 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_+_184055240 | 0.59 |
ENST00000383847.2
|
FAM131A
|
family with sequence similarity 131, member A |
| chr7_-_138363824 | 0.58 |
ENST00000419765.3
|
SVOPL
|
SVOP-like |
| chr12_+_124155652 | 0.58 |
ENST00000426174.2
ENST00000303372.5 |
TCTN2
|
tectonic family member 2 |
| chr14_-_106069247 | 0.57 |
ENST00000479229.1
|
RP11-731F5.1
|
RP11-731F5.1 |
| chr19_+_36236491 | 0.57 |
ENST00000591949.1
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr19_+_36236514 | 0.56 |
ENST00000222266.2
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr20_+_58179582 | 0.55 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr19_-_48823332 | 0.54 |
ENST00000315396.7
|
CCDC114
|
coiled-coil domain containing 114 |
| chr11_+_60197069 | 0.53 |
ENST00000528905.1
ENST00000528093.1 |
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr8_-_12668962 | 0.52 |
ENST00000534827.1
|
RP11-252C15.1
|
RP11-252C15.1 |
| chr20_+_32254286 | 0.50 |
ENST00000330271.4
|
ACTL10
|
actin-like 10 |
| chr5_+_98109322 | 0.48 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr2_+_162272605 | 0.48 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr1_+_210406121 | 0.48 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr21_-_47743719 | 0.47 |
ENST00000397680.1
ENST00000445935.1 ENST00000397685.4 ENST00000397682.3 ENST00000291691.7 |
C21orf58
|
chromosome 21 open reading frame 58 |
| chr10_-_102279586 | 0.46 |
ENST00000370345.3
ENST00000451524.1 ENST00000370329.5 |
SEC31B
|
SEC31 homolog B (S. cerevisiae) |
| chr10_-_13523073 | 0.45 |
ENST00000440282.1
|
BEND7
|
BEN domain containing 7 |
| chr16_+_4838412 | 0.45 |
ENST00000589327.1
|
SMIM22
|
small integral membrane protein 22 |
| chr6_-_161695042 | 0.44 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr19_+_17337473 | 0.44 |
ENST00000598068.1
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr7_+_120591170 | 0.43 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family, member 3 |
| chr10_+_94590910 | 0.41 |
ENST00000371547.4
|
EXOC6
|
exocyst complex component 6 |
| chr3_+_125694347 | 0.40 |
ENST00000505382.1
ENST00000511082.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr14_-_23285069 | 0.39 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr6_-_31648150 | 0.39 |
ENST00000375858.3
ENST00000383237.4 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr16_-_18468926 | 0.39 |
ENST00000545114.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr14_-_23285011 | 0.39 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr6_+_90272027 | 0.38 |
ENST00000522441.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr6_-_56707943 | 0.38 |
ENST00000370769.4
ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST
|
dystonin |
| chr2_-_96874553 | 0.37 |
ENST00000337288.5
ENST00000443962.1 |
STARD7
|
StAR-related lipid transfer (START) domain containing 7 |
| chr3_-_187454281 | 0.37 |
ENST00000232014.4
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr19_+_36602104 | 0.37 |
ENST00000585332.1
ENST00000262637.4 |
OVOL3
|
ovo-like zinc finger 3 |
| chr15_-_49255632 | 0.37 |
ENST00000332408.4
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4 |
| chr16_+_30212378 | 0.36 |
ENST00000569485.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr1_-_173638976 | 0.36 |
ENST00000333279.2
|
ANKRD45
|
ankyrin repeat domain 45 |
| chr6_-_161695074 | 0.36 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr3_-_190580404 | 0.35 |
ENST00000442080.1
|
GMNC
|
geminin coiled-coil domain containing |
| chr12_+_50017327 | 0.34 |
ENST00000261897.1
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr19_-_17571722 | 0.34 |
ENST00000301944.2
|
NXNL1
|
nucleoredoxin-like 1 |
| chr21_-_34185944 | 0.33 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr18_+_48494361 | 0.32 |
ENST00000588577.1
ENST00000269466.3 ENST00000591429.1 ENST00000452201.2 |
ELAC1
SMAD4
|
elaC ribonuclease Z 1 SMAD family member 4 |
| chr16_+_4838393 | 0.31 |
ENST00000589721.1
|
SMIM22
|
small integral membrane protein 22 |
| chr15_-_27184664 | 0.30 |
ENST00000541819.2
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr11_-_94964354 | 0.30 |
ENST00000536441.1
|
SESN3
|
sestrin 3 |
| chr1_+_45965725 | 0.29 |
ENST00000401061.4
|
MMACHC
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr17_-_9929581 | 0.29 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr15_-_90645679 | 0.29 |
ENST00000539790.1
ENST00000559482.1 ENST00000330062.3 |
IDH2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr1_+_164528866 | 0.29 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr16_+_29472707 | 0.28 |
ENST00000565290.1
|
SULT1A4
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr1_+_177140633 | 0.28 |
ENST00000361539.4
|
BRINP2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr7_+_120590803 | 0.27 |
ENST00000315870.5
ENST00000339121.5 ENST00000445699.1 |
ING3
|
inhibitor of growth family, member 3 |
| chr6_-_36842784 | 0.27 |
ENST00000373699.5
|
PPIL1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chrX_+_129473916 | 0.27 |
ENST00000545805.1
ENST00000543953.1 ENST00000218197.5 |
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr5_+_133451254 | 0.27 |
ENST00000517851.1
ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr11_-_85397167 | 0.27 |
ENST00000316398.3
|
CCDC89
|
coiled-coil domain containing 89 |
| chr17_-_79995553 | 0.27 |
ENST00000581584.1
ENST00000577712.1 ENST00000582900.1 ENST00000579155.1 ENST00000306869.2 |
DCXR
|
dicarbonyl/L-xylulose reductase |
| chr3_-_93781750 | 0.26 |
ENST00000314636.2
|
DHFRL1
|
dihydrofolate reductase-like 1 |
| chr19_-_45681482 | 0.26 |
ENST00000592647.1
ENST00000006275.4 ENST00000588062.1 ENST00000585934.1 |
TRAPPC6A
|
trafficking protein particle complex 6A |
| chr6_+_32821924 | 0.26 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr19_-_46389359 | 0.26 |
ENST00000302165.3
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1 |
| chr9_-_123239632 | 0.26 |
ENST00000416449.1
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr12_+_50017184 | 0.26 |
ENST00000548825.2
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr1_+_15272271 | 0.26 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr1_+_204839959 | 0.25 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chr18_-_30050395 | 0.25 |
ENST00000269209.6
ENST00000399218.4 |
GAREM
|
GRB2 associated, regulator of MAPK1 |
| chr16_+_15039742 | 0.24 |
ENST00000537062.1
|
NPIPA1
|
nuclear pore complex interacting protein family, member A1 |
| chr19_+_58258164 | 0.24 |
ENST00000317178.5
|
ZNF776
|
zinc finger protein 776 |
| chr13_-_67802549 | 0.24 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr12_+_109273806 | 0.24 |
ENST00000228476.3
ENST00000547768.1 |
DAO
|
D-amino-acid oxidase |
| chr7_+_123488124 | 0.24 |
ENST00000476325.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr22_+_45072958 | 0.23 |
ENST00000403581.1
|
PRR5
|
proline rich 5 (renal) |
| chr6_-_56708459 | 0.23 |
ENST00000370788.2
|
DST
|
dystonin |
| chr22_+_45072925 | 0.23 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr11_+_35684288 | 0.23 |
ENST00000299413.5
|
TRIM44
|
tripartite motif containing 44 |
| chr19_-_10687907 | 0.23 |
ENST00000589348.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr11_+_119056178 | 0.23 |
ENST00000525131.1
ENST00000531114.1 ENST00000355547.5 ENST00000322712.4 |
PDZD3
|
PDZ domain containing 3 |
| chr11_-_68780824 | 0.23 |
ENST00000441623.1
ENST00000309099.6 |
MRGPRF
|
MAS-related GPR, member F |
| chr19_+_55477711 | 0.23 |
ENST00000448584.2
ENST00000537859.1 ENST00000585500.1 ENST00000427260.2 ENST00000538819.1 ENST00000263437.6 |
NLRP2
|
NLR family, pyrin domain containing 2 |
| chr2_+_61372226 | 0.23 |
ENST00000426997.1
|
C2orf74
|
chromosome 2 open reading frame 74 |
| chr3_+_152017924 | 0.23 |
ENST00000465907.2
ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr10_+_71211212 | 0.23 |
ENST00000373290.2
|
TSPAN15
|
tetraspanin 15 |
| chr16_+_50775948 | 0.22 |
ENST00000569681.1
ENST00000569418.1 ENST00000540145.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr1_+_182419261 | 0.22 |
ENST00000294854.8
ENST00000542961.1 |
RGSL1
|
regulator of G-protein signaling like 1 |
| chr4_-_156875003 | 0.22 |
ENST00000433477.3
|
CTSO
|
cathepsin O |
| chr1_+_164529004 | 0.22 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr1_+_15943995 | 0.22 |
ENST00000480945.1
|
DDI2
|
DNA-damage inducible 1 homolog 2 (S. cerevisiae) |
| chr19_+_7069690 | 0.22 |
ENST00000439035.2
|
ZNF557
|
zinc finger protein 557 |
| chr4_+_154125565 | 0.21 |
ENST00000338700.5
|
TRIM2
|
tripartite motif containing 2 |
| chr2_-_158732340 | 0.21 |
ENST00000539637.1
ENST00000413751.1 ENST00000434821.1 ENST00000424669.1 |
ACVR1
|
activin A receptor, type I |
| chr20_+_52105495 | 0.21 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr1_-_10856694 | 0.21 |
ENST00000377022.3
ENST00000344008.5 |
CASZ1
|
castor zinc finger 1 |
| chr3_-_172312460 | 0.21 |
ENST00000418839.2
|
RP11-408H1.3
|
RP11-408H1.3 |
| chr7_+_107110488 | 0.21 |
ENST00000304402.4
|
GPR22
|
G protein-coupled receptor 22 |
| chr2_+_47630108 | 0.20 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr4_+_76995855 | 0.19 |
ENST00000355810.4
ENST00000349321.3 |
ART3
|
ADP-ribosyltransferase 3 |
| chrX_-_48056199 | 0.19 |
ENST00000311798.1
ENST00000347757.1 |
SSX5
|
synovial sarcoma, X breakpoint 5 |
| chr2_-_45236540 | 0.19 |
ENST00000303077.6
|
SIX2
|
SIX homeobox 2 |
| chr6_+_44215603 | 0.19 |
ENST00000371554.1
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr13_+_113623509 | 0.19 |
ENST00000535094.2
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr11_-_33774944 | 0.19 |
ENST00000532057.1
ENST00000531080.1 |
FBXO3
|
F-box protein 3 |
| chr22_+_41865109 | 0.18 |
ENST00000216254.4
ENST00000396512.3 |
ACO2
|
aconitase 2, mitochondrial |
| chr1_-_13007420 | 0.18 |
ENST00000376189.1
|
PRAMEF6
|
PRAME family member 6 |
| chr1_-_23886285 | 0.18 |
ENST00000374561.5
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr10_+_88516396 | 0.17 |
ENST00000372037.3
|
BMPR1A
|
bone morphogenetic protein receptor, type IA |
| chr7_+_142364193 | 0.17 |
ENST00000390397.2
|
TRBV24-1
|
T cell receptor beta variable 24-1 |
| chr11_-_62420757 | 0.17 |
ENST00000330574.2
|
INTS5
|
integrator complex subunit 5 |
| chr1_+_39796810 | 0.17 |
ENST00000289893.4
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr1_-_49242553 | 0.17 |
ENST00000371833.3
|
BEND5
|
BEN domain containing 5 |
| chr5_+_56111361 | 0.17 |
ENST00000399503.3
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr1_-_13005246 | 0.17 |
ENST00000415464.2
|
PRAMEF6
|
PRAME family member 6 |
| chr7_+_150758304 | 0.17 |
ENST00000482950.1
ENST00000463414.1 ENST00000310317.5 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr6_-_116381918 | 0.17 |
ENST00000606080.1
|
FRK
|
fyn-related kinase |
| chr9_-_110540419 | 0.17 |
ENST00000398726.3
|
AL162389.1
|
Uncharacterized protein |
| chr14_+_77228532 | 0.16 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr6_+_107077435 | 0.16 |
ENST00000369046.4
|
QRSL1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr19_-_35719609 | 0.16 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187, member B |
| chr12_+_53835508 | 0.16 |
ENST00000551003.1
ENST00000549068.1 ENST00000549740.1 ENST00000546581.1 ENST00000549581.1 ENST00000541275.1 |
PRR13
PCBP2
|
proline rich 13 poly(rC) binding protein 2 |
| chr7_+_73106926 | 0.16 |
ENST00000453316.1
|
WBSCR22
|
Williams Beuren syndrome chromosome region 22 |
| chr17_-_7218631 | 0.16 |
ENST00000577040.2
ENST00000389167.5 ENST00000391950.3 |
GPS2
|
G protein pathway suppressor 2 |
| chr19_+_57862622 | 0.15 |
ENST00000391705.3
ENST00000443917.2 ENST00000598744.1 |
ZNF304
|
zinc finger protein 304 |
| chr16_+_19183671 | 0.15 |
ENST00000562711.2
|
SYT17
|
synaptotagmin XVII |
| chr9_+_35538616 | 0.15 |
ENST00000455600.1
|
RUSC2
|
RUN and SH3 domain containing 2 |
| chr1_-_19615744 | 0.15 |
ENST00000361640.4
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr1_+_13359819 | 0.15 |
ENST00000376168.1
|
PRAMEF5
|
PRAME family member 5 |
| chr14_+_78870030 | 0.15 |
ENST00000553631.1
ENST00000554719.1 |
NRXN3
|
neurexin 3 |
| chr17_+_36858694 | 0.15 |
ENST00000563897.1
|
CTB-58E17.1
|
CTB-58E17.1 |
| chr1_+_109102652 | 0.15 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr1_+_14075903 | 0.15 |
ENST00000343137.4
ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr20_-_35274548 | 0.15 |
ENST00000262866.4
|
SLA2
|
Src-like-adaptor 2 |
| chr12_+_53835383 | 0.15 |
ENST00000429243.2
|
PRR13
|
proline rich 13 |
| chr14_+_105957402 | 0.14 |
ENST00000421892.1
ENST00000334656.7 ENST00000451719.1 ENST00000392522.3 ENST00000392523.4 ENST00000354560.6 ENST00000450383.1 |
C14orf80
|
chromosome 14 open reading frame 80 |
| chr16_-_75301886 | 0.14 |
ENST00000393422.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr9_-_70488865 | 0.14 |
ENST00000377392.5
|
CBWD5
|
COBW domain containing 5 |
| chr5_-_137071756 | 0.14 |
ENST00000394937.3
ENST00000309755.4 |
KLHL3
|
kelch-like family member 3 |
| chr17_+_3100813 | 0.14 |
ENST00000381951.1
|
OR1A2
|
olfactory receptor, family 1, subfamily A, member 2 |
| chr8_+_62200509 | 0.14 |
ENST00000519846.1
ENST00000518592.1 ENST00000325897.4 |
CLVS1
|
clavesin 1 |
| chr12_+_53835539 | 0.14 |
ENST00000547368.1
ENST00000379786.4 ENST00000551945.1 |
PRR13
|
proline rich 13 |
| chr16_+_30675654 | 0.14 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr2_-_9563575 | 0.13 |
ENST00000488451.1
ENST00000238091.4 ENST00000355346.4 |
ITGB1BP1
|
integrin beta 1 binding protein 1 |
| chr7_-_99332719 | 0.13 |
ENST00000336374.2
|
CYP3A7
|
cytochrome P450, family 3, subfamily A, polypeptide 7 |
| chr19_-_44124019 | 0.13 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr2_+_242295658 | 0.13 |
ENST00000264042.3
ENST00000545004.1 ENST00000373287.4 |
FARP2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr12_+_48513009 | 0.13 |
ENST00000359794.5
ENST00000551339.1 ENST00000395233.2 ENST00000548345.1 |
PFKM
|
phosphofructokinase, muscle |
| chr10_+_45455207 | 0.13 |
ENST00000334940.6
ENST00000374417.2 ENST00000340258.5 ENST00000427758.1 |
RASSF4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr21_-_43430440 | 0.13 |
ENST00000398505.3
ENST00000310826.5 ENST00000449949.1 ENST00000398499.1 ENST00000398497.2 ENST00000398511.3 |
ZBTB21
|
zinc finger and BTB domain containing 21 |
| chr16_-_4838255 | 0.13 |
ENST00000591624.1
ENST00000396693.5 |
SEPT12
|
septin 12 |
| chr1_-_182360918 | 0.13 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr16_+_50775971 | 0.13 |
ENST00000311559.9
ENST00000564326.1 ENST00000566206.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr19_+_11649532 | 0.13 |
ENST00000252456.2
ENST00000592923.1 ENST00000535659.2 |
CNN1
|
calponin 1, basic, smooth muscle |
| chr19_+_11455900 | 0.13 |
ENST00000588790.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr5_+_37379314 | 0.13 |
ENST00000265107.4
ENST00000504564.1 |
WDR70
|
WD repeat domain 70 |
| chr11_+_61717336 | 0.12 |
ENST00000378042.3
|
BEST1
|
bestrophin 1 |
| chr1_-_13002348 | 0.12 |
ENST00000355096.2
|
PRAMEF6
|
PRAME family member 6 |
| chr2_+_202122703 | 0.12 |
ENST00000447616.1
ENST00000358485.4 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr1_+_158325684 | 0.12 |
ENST00000368162.2
|
CD1E
|
CD1e molecule |
| chr17_+_61699766 | 0.12 |
ENST00000579585.1
ENST00000584573.1 ENST00000361733.3 ENST00000361357.3 |
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr17_-_56595196 | 0.12 |
ENST00000579921.1
ENST00000579925.1 ENST00000323456.5 |
MTMR4
|
myotubularin related protein 4 |
| chr2_+_26915584 | 0.12 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3 |
| chr13_+_111806055 | 0.12 |
ENST00000218789.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr20_+_47835884 | 0.12 |
ENST00000371764.4
|
DDX27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr1_-_182361327 | 0.11 |
ENST00000331872.6
ENST00000311223.5 |
GLUL
|
glutamate-ammonia ligase |
| chr14_-_64108125 | 0.11 |
ENST00000267522.3
|
WDR89
|
WD repeat domain 89 |
| chr12_+_48513570 | 0.11 |
ENST00000551804.1
|
PFKM
|
phosphofructokinase, muscle |
| chr11_-_5173599 | 0.11 |
ENST00000328942.1
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1 |
| chr12_-_70093190 | 0.11 |
ENST00000330891.5
|
BEST3
|
bestrophin 3 |
| chr1_+_226736446 | 0.11 |
ENST00000366788.3
ENST00000366789.4 |
C1orf95
|
chromosome 1 open reading frame 95 |
| chr3_+_152017360 | 0.11 |
ENST00000485910.1
ENST00000463374.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr4_+_160188889 | 0.11 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr10_+_24498060 | 0.11 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr2_+_69201705 | 0.11 |
ENST00000377938.2
|
GKN1
|
gastrokine 1 |
| chr10_+_24528108 | 0.10 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr9_-_115819039 | 0.10 |
ENST00000555206.1
|
ZFP37
|
ZFP37 zinc finger protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of GFI1B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.2 | 0.7 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 1.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 3.0 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 0.8 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.4 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 0.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.8 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 1.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.3 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.1 | 0.4 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.6 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.3 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.1 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.2 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.1 | 0.2 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 4.8 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.2 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.2 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 0.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.3 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 0.2 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 1.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.2 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.2 | GO:0048378 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) |
| 0.0 | 0.2 | GO:0044467 | glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.0 | 0.8 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:0052148 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.2 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.0 | 0.1 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.0 | 0.8 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 1.2 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.1 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 1.2 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.5 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.1 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.1 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 0.6 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 2.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.8 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.2 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.3 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.4 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.2 | 0.7 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 1.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.3 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.2 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 1.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.2 | GO:0032181 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.1 | 0.2 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 0.2 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.4 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 0.3 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 0.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 1.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.4 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |