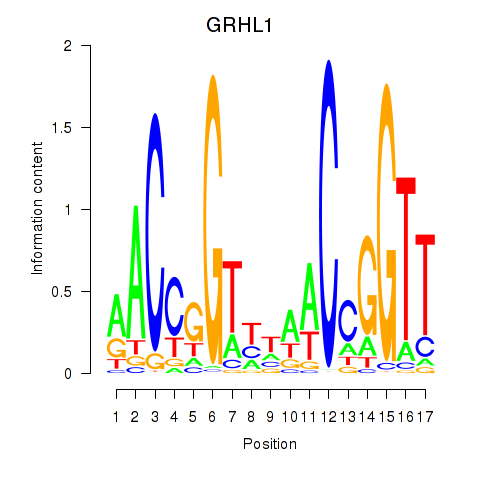
|
chr13_-_20806440
Show fit
|
7.35 |
ENST00000400066.3
ENST00000400065.3
ENST00000356192.6
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr1_-_153113927
Show fit
|
5.80 |
ENST00000368752.4
|
SPRR2B
|
small proline-rich protein 2B
|
|
chr7_-_102283238
Show fit
|
2.78 |
ENST00000340457.8
|
UPK3BL
|
uroplakin 3B-like
|
|
chr7_-_102184083
Show fit
|
2.72 |
ENST00000379357.5
|
POLR2J3
|
polymerase (RNA) II (DNA directed) polypeptide J3
|
|
chr1_-_24469602
Show fit
|
2.41 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr11_+_706595
Show fit
|
2.19 |
ENST00000531348.1
ENST00000530636.1
|
EPS8L2
|
EPS8-like 2
|
|
chr7_+_76139925
Show fit
|
2.02 |
ENST00000394849.1
|
UPK3B
|
uroplakin 3B
|
|
chr9_+_5450503
Show fit
|
1.99 |
ENST00000381573.4
ENST00000381577.3
|
CD274
|
CD274 molecule
|
|
chr20_-_54967187
Show fit
|
1.97 |
ENST00000422322.1
ENST00000371356.2
ENST00000451915.1
ENST00000347343.2
ENST00000395911.1
ENST00000395907.1
ENST00000441357.1
ENST00000456249.1
ENST00000420474.1
ENST00000395909.4
ENST00000395914.1
ENST00000312783.6
ENST00000395915.3
ENST00000395913.3
|
AURKA
|
aurora kinase A
|
|
chr14_-_80677815
Show fit
|
1.90 |
ENST00000557125.1
ENST00000555750.1
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr7_+_76139833
Show fit
|
1.88 |
ENST00000257632.5
|
UPK3B
|
uroplakin 3B
|
|
chr3_+_101568349
Show fit
|
1.78 |
ENST00000326151.5
ENST00000326172.5
|
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr2_-_85641162
Show fit
|
1.75 |
ENST00000447219.2
ENST00000409670.1
ENST00000409724.1
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr2_+_113816215
Show fit
|
1.74 |
ENST00000346807.3
|
IL36RN
|
interleukin 36 receptor antagonist
|
|
chr7_-_80551671
Show fit
|
1.73 |
ENST00000419255.2
ENST00000544525.1
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr11_-_67442196
Show fit
|
1.67 |
ENST00000525827.1
|
ALDH3B2
|
aldehyde dehydrogenase 3 family, member B2
|
|
chr17_+_9728828
Show fit
|
1.51 |
ENST00000262441.5
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr1_-_201368707
Show fit
|
1.49 |
ENST00000391967.2
|
LAD1
|
ladinin 1
|
|
chr2_+_95940220
Show fit
|
1.48 |
ENST00000542147.1
|
PROM2
|
prominin 2
|
|
chr1_+_155023757
Show fit
|
1.45 |
ENST00000356955.2
ENST00000449910.2
ENST00000359280.4
ENST00000360674.4
ENST00000368412.3
ENST00000355956.2
ENST00000368410.2
ENST00000271836.6
ENST00000368413.1
ENST00000531455.1
ENST00000447332.3
|
ADAM15
|
ADAM metallopeptidase domain 15
|
|
chr10_+_105036909
Show fit
|
1.44 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr1_-_201368653
Show fit
|
1.42 |
ENST00000367313.3
|
LAD1
|
ladinin 1
|
|
chr14_-_80677613
Show fit
|
1.40 |
ENST00000556811.1
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr8_-_130799134
Show fit
|
1.35 |
ENST00000276708.4
|
GSDMC
|
gasdermin C
|
|
chr2_+_95940186
Show fit
|
1.33 |
ENST00000403131.2
ENST00000317668.4
ENST00000317620.9
|
PROM2
|
prominin 2
|
|
chr11_-_67442079
Show fit
|
1.31 |
ENST00000349015.3
|
ALDH3B2
|
aldehyde dehydrogenase 3 family, member B2
|
|
chr1_+_35247859
Show fit
|
1.30 |
ENST00000373362.3
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr19_+_19496728
Show fit
|
1.26 |
ENST00000537887.1
ENST00000417582.2
|
GATAD2A
|
GATA zinc finger domain containing 2A
|
|
chr12_+_123874589
Show fit
|
1.05 |
ENST00000437502.1
|
SETD8
|
SET domain containing (lysine methyltransferase) 8
|
|
chr7_+_76139741
Show fit
|
1.03 |
ENST00000334348.3
ENST00000419923.2
ENST00000448265.3
ENST00000443097.2
|
UPK3B
|
uroplakin 3B
|
|
chr1_-_28384598
Show fit
|
1.03 |
ENST00000373864.1
|
EYA3
|
eyes absent homolog 3 (Drosophila)
|
|
chr6_+_32121218
Show fit
|
1.03 |
ENST00000414204.1
ENST00000361568.2
ENST00000395523.1
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr20_-_61002584
Show fit
|
1.00 |
ENST00000252998.1
|
RBBP8NL
|
RBBP8 N-terminal like
|
|
chr11_+_119039414
Show fit
|
0.96 |
ENST00000409991.1
ENST00000292199.2
ENST00000409265.4
ENST00000409109.1
|
NLRX1
|
NLR family member X1
|
|
chr19_-_15590306
Show fit
|
0.95 |
ENST00000292609.4
|
PGLYRP2
|
peptidoglycan recognition protein 2
|
|
chr18_-_35145728
Show fit
|
0.92 |
ENST00000361795.5
ENST00000603232.1
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr19_-_17356697
Show fit
|
0.92 |
ENST00000291442.3
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6
|
|
chr9_+_77703414
Show fit
|
0.85 |
ENST00000346234.6
|
OSTF1
|
osteoclast stimulating factor 1
|
|
chr3_-_49142504
Show fit
|
0.78 |
ENST00000306125.6
ENST00000420147.2
|
QARS
|
glutaminyl-tRNA synthetase
|
|
chr5_-_132299313
Show fit
|
0.78 |
ENST00000265343.5
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr20_-_25320367
Show fit
|
0.69 |
ENST00000450393.1
ENST00000491682.1
|
ABHD12
|
abhydrolase domain containing 12
|
|
chr19_+_19496624
Show fit
|
0.65 |
ENST00000494516.2
ENST00000360315.3
ENST00000252577.5
|
GATAD2A
|
GATA zinc finger domain containing 2A
|
|
chr3_-_49142178
Show fit
|
0.64 |
ENST00000452739.1
ENST00000414533.1
ENST00000417025.1
|
QARS
|
glutaminyl-tRNA synthetase
|
|
chr18_+_9475001
Show fit
|
0.63 |
ENST00000019317.4
|
RALBP1
|
ralA binding protein 1
|
|
chr9_-_77703115
Show fit
|
0.62 |
ENST00000361092.4
ENST00000376808.4
|
NMRK1
|
nicotinamide riboside kinase 1
|
|
chr19_+_10736183
Show fit
|
0.60 |
ENST00000590857.1
ENST00000588688.1
ENST00000586078.1
|
SLC44A2
|
solute carrier family 44 (choline transporter), member 2
|
|
chr11_-_69590101
Show fit
|
0.50 |
ENST00000168712.1
|
FGF4
|
fibroblast growth factor 4
|
|
chr2_-_209118974
Show fit
|
0.50 |
ENST00000415913.1
ENST00000415282.1
ENST00000446179.1
|
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble
|
|
chr6_-_31107127
Show fit
|
0.49 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2
|
|
chr7_-_138794081
Show fit
|
0.46 |
ENST00000464606.1
|
ZC3HAV1
|
zinc finger CCCH-type, antiviral 1
|
|
chr1_-_51425772
Show fit
|
0.45 |
ENST00000371778.4
|
FAF1
|
Fas (TNFRSF6) associated factor 1
|
|
chr12_+_18414446
Show fit
|
0.40 |
ENST00000433979.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma
|
|
chr4_+_156588249
Show fit
|
0.39 |
ENST00000393832.3
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr14_-_23540826
Show fit
|
0.36 |
ENST00000357481.2
|
ACIN1
|
apoptotic chromatin condensation inducer 1
|
|
chr19_+_58038683
Show fit
|
0.35 |
ENST00000240719.3
ENST00000376233.3
ENST00000594943.1
ENST00000602149.1
|
ZNF549
|
zinc finger protein 549
|
|
chr1_+_44440575
Show fit
|
0.31 |
ENST00000532642.1
ENST00000236067.4
ENST00000471859.2
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr18_-_35145981
Show fit
|
0.30 |
ENST00000420428.2
ENST00000412753.1
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr5_-_132299290
Show fit
|
0.29 |
ENST00000378595.3
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr4_-_99850243
Show fit
|
0.24 |
ENST00000280892.6
ENST00000511644.1
ENST00000504432.1
ENST00000505992.1
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr18_-_35145689
Show fit
|
0.20 |
ENST00000591287.1
ENST00000601019.1
ENST00000601392.1
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr1_+_206643806
Show fit
|
0.18 |
ENST00000537984.1
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon
|
|
chr6_-_43027105
Show fit
|
0.13 |
ENST00000230413.5
ENST00000487429.1
ENST00000489623.1
ENST00000468957.1
|
MRPL2
|
mitochondrial ribosomal protein L2
|
|
chrX_+_146993449
Show fit
|
0.13 |
ENST00000218200.8
ENST00000370471.3
ENST00000370477.1
|
FMR1
|
fragile X mental retardation 1
|
|
chr18_-_35145593
Show fit
|
0.11 |
ENST00000334919.5
ENST00000591282.1
ENST00000588597.1
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr1_-_16539094
Show fit
|
0.11 |
ENST00000270747.3
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19
|
|
chr9_+_71736177
Show fit
|
0.07 |
ENST00000606364.1
ENST00000453658.2
|
TJP2
|
tight junction protein 2
|
|
chr1_-_234667504
Show fit
|
0.06 |
ENST00000421207.1
ENST00000435574.1
|
RP5-855F14.1
|
RP5-855F14.1
|
|
chr1_-_36863481
Show fit
|
0.04 |
ENST00000315732.2
|
LSM10
|
LSM10, U7 small nuclear RNA associated
|
|
chr5_+_134303591
Show fit
|
0.03 |
ENST00000282611.6
|
CATSPER3
|
cation channel, sperm associated 3
|
|
chr9_+_130565487
Show fit
|
0.00 |
ENST00000373225.3
ENST00000431857.1
|
FPGS
|
folylpolyglutamate synthase
|