Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
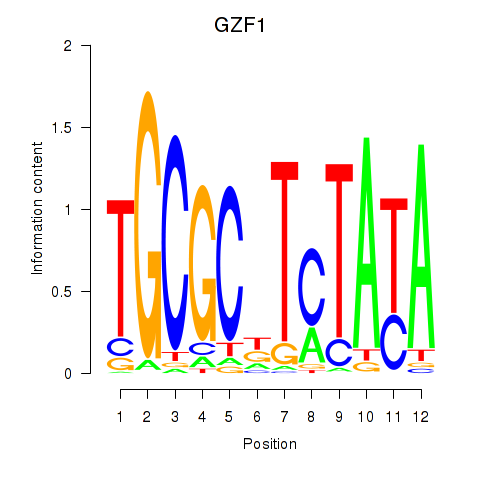
Results for GZF1
Z-value: 0.38
Transcription factors associated with GZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GZF1
|
ENSG00000125812.11 | GDNF inducible zinc finger protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GZF1 | hg19_v2_chr20_+_23342783_23342957 | 0.38 | 3.9e-02 | Click! |
Activity profile of GZF1 motif
Sorted Z-values of GZF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_101918153 | 2.01 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr20_+_42086525 | 0.97 |
ENST00000244020.3
|
SRSF6
|
serine/arginine-rich splicing factor 6 |
| chr1_+_79115503 | 0.96 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr9_-_124976185 | 0.79 |
ENST00000464484.2
|
LHX6
|
LIM homeobox 6 |
| chr9_-_124976154 | 0.79 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chr14_-_92413727 | 0.68 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr12_+_104458235 | 0.65 |
ENST00000229330.4
|
HCFC2
|
host cell factor C2 |
| chr3_-_114035026 | 0.64 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr9_+_96338647 | 0.62 |
ENST00000359246.4
|
PHF2
|
PHD finger protein 2 |
| chr18_-_53303123 | 0.61 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr9_+_96338860 | 0.51 |
ENST00000375376.4
|
PHF2
|
PHD finger protein 2 |
| chr1_-_32801825 | 0.50 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr3_-_79816965 | 0.46 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr1_-_144995074 | 0.40 |
ENST00000534536.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr19_-_36545128 | 0.38 |
ENST00000538849.1
|
THAP8
|
THAP domain containing 8 |
| chr3_+_30647994 | 0.36 |
ENST00000295754.5
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr14_-_92413353 | 0.36 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr14_-_21979428 | 0.34 |
ENST00000538267.1
ENST00000298717.4 |
METTL3
|
methyltransferase like 3 |
| chr1_-_144994909 | 0.32 |
ENST00000369347.4
ENST00000369354.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr1_-_11107280 | 0.28 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr15_+_44580955 | 0.25 |
ENST00000345795.2
ENST00000360824.3 |
CASC4
|
cancer susceptibility candidate 4 |
| chr1_-_144995002 | 0.24 |
ENST00000369356.4
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr20_-_31331212 | 0.24 |
ENST00000474815.2
ENST00000446419.2 |
COMMD7
|
COMM domain containing 7 |
| chr14_-_53619816 | 0.24 |
ENST00000323669.5
ENST00000395606.1 ENST00000357758.3 |
DDHD1
|
DDHD domain containing 1 |
| chr3_+_30648066 | 0.22 |
ENST00000359013.4
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr17_-_70588936 | 0.21 |
ENST00000453722.2
|
LINC00511
|
long intergenic non-protein coding RNA 511 |
| chr1_+_41174988 | 0.20 |
ENST00000372652.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chrX_+_102883620 | 0.19 |
ENST00000372626.3
|
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr16_-_30582888 | 0.18 |
ENST00000563707.1
ENST00000567855.1 |
ZNF688
|
zinc finger protein 688 |
| chr10_+_60028818 | 0.18 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr16_+_67876180 | 0.17 |
ENST00000303596.1
|
THAP11
|
THAP domain containing 11 |
| chr8_+_23430157 | 0.17 |
ENST00000399967.3
|
FP15737
|
FP15737 |
| chr2_-_27603582 | 0.14 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr10_+_89264625 | 0.13 |
ENST00000371996.4
ENST00000371994.4 |
MINPP1
|
multiple inositol-polyphosphate phosphatase 1 |
| chr10_+_14880287 | 0.12 |
ENST00000437161.2
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr9_-_14308004 | 0.11 |
ENST00000493697.1
|
NFIB
|
nuclear factor I/B |
| chr1_-_144994840 | 0.11 |
ENST00000369351.3
ENST00000369349.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr4_-_89619386 | 0.11 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr19_+_36157715 | 0.10 |
ENST00000379013.2
ENST00000222275.2 |
UPK1A
|
uroplakin 1A |
| chr3_+_51705222 | 0.10 |
ENST00000457573.1
ENST00000341333.5 ENST00000412249.1 ENST00000425781.1 ENST00000415259.1 ENST00000395057.1 ENST00000416589.1 |
TEX264
|
testis expressed 264 |
| chr1_-_2461684 | 0.08 |
ENST00000378453.3
|
HES5
|
hes family bHLH transcription factor 5 |
| chr5_+_92919043 | 0.07 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr10_-_126849588 | 0.05 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr16_-_46797149 | 0.05 |
ENST00000536476.1
|
MYLK3
|
myosin light chain kinase 3 |
| chr22_-_43355858 | 0.05 |
ENST00000402229.1
ENST00000407585.1 ENST00000453079.1 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr10_-_126849068 | 0.03 |
ENST00000494626.2
ENST00000337195.5 |
CTBP2
|
C-terminal binding protein 2 |
| chr10_-_14880566 | 0.03 |
ENST00000378442.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr14_+_22293618 | 0.03 |
ENST00000390432.2
|
TRAV10
|
T cell receptor alpha variable 10 |
| chr16_-_70285797 | 0.02 |
ENST00000435634.1
|
EXOSC6
|
exosome component 6 |
| chr3_-_170588163 | 0.02 |
ENST00000295830.8
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr19_-_52150053 | 0.02 |
ENST00000599649.1
ENST00000429354.3 ENST00000360844.6 ENST00000222107.4 |
SIGLEC5
SIGLEC14
SIGLEC5
|
SIGLEC5 sialic acid binding Ig-like lectin 14 sialic acid binding Ig-like lectin 5 |
| chr4_-_147443043 | 0.02 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr19_-_10426663 | 0.01 |
ENST00000541276.1
ENST00000393708.3 ENST00000494368.1 |
FDX1L
|
ferredoxin 1-like |
| chr7_+_102004322 | 0.01 |
ENST00000496391.1
|
PRKRIP1
|
PRKR interacting protein 1 (IL11 inducible) |
Network of associatons between targets according to the STRING database.
First level regulatory network of GZF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.2 | 0.6 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.2 | 0.5 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.1 | 1.6 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 1.0 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 1.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.3 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.6 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.3 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.1 | 1.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 1.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |