Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for HDX
Z-value: 0.29
Transcription factors associated with HDX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HDX
|
ENSG00000165259.9 | highly divergent homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HDX | hg19_v2_chrX_-_83757399_83757487 | 0.39 | 3.5e-02 | Click! |
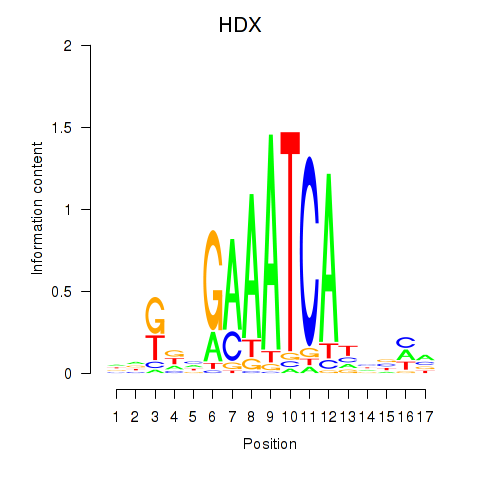
Activity profile of HDX motif
Sorted Z-values of HDX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_113594279 | 0.55 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr2_+_128403720 | 0.35 |
ENST00000272644.3
|
GPR17
|
G protein-coupled receptor 17 |
| chr1_+_84630053 | 0.33 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr2_+_128403439 | 0.33 |
ENST00000544369.1
|
GPR17
|
G protein-coupled receptor 17 |
| chr7_-_69062391 | 0.31 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr18_+_29027696 | 0.30 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr2_+_113735575 | 0.26 |
ENST00000376489.2
ENST00000259205.4 |
IL36G
|
interleukin 36, gamma |
| chr5_-_35230434 | 0.24 |
ENST00000504500.1
|
PRLR
|
prolactin receptor |
| chr12_+_101869096 | 0.21 |
ENST00000551346.1
|
SPIC
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr5_-_41213607 | 0.19 |
ENST00000337836.5
ENST00000433294.1 |
C6
|
complement component 6 |
| chr19_-_41934635 | 0.17 |
ENST00000321702.2
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr1_+_117602925 | 0.16 |
ENST00000369466.4
|
TTF2
|
transcription termination factor, RNA polymerase II |
| chr5_+_60933634 | 0.15 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr3_-_165555200 | 0.12 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr12_+_10460549 | 0.12 |
ENST00000543420.1
ENST00000543777.1 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr7_-_121944491 | 0.11 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr10_-_14372870 | 0.11 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr1_-_169680745 | 0.10 |
ENST00000236147.4
|
SELL
|
selectin L |
| chr5_-_35230649 | 0.09 |
ENST00000382002.5
|
PRLR
|
prolactin receptor |
| chr7_-_135412925 | 0.08 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr11_-_66675371 | 0.08 |
ENST00000393955.2
|
PC
|
pyruvate carboxylase |
| chr12_+_54422142 | 0.08 |
ENST00000243108.4
|
HOXC6
|
homeobox C6 |
| chr22_+_40440804 | 0.08 |
ENST00000441751.1
ENST00000301923.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr7_+_99954224 | 0.08 |
ENST00000608825.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr5_-_35230771 | 0.08 |
ENST00000342362.5
|
PRLR
|
prolactin receptor |
| chr6_+_144471643 | 0.08 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chr12_+_10460417 | 0.07 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr2_+_169926047 | 0.06 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr1_-_11024258 | 0.06 |
ENST00000418570.2
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr5_-_135290705 | 0.05 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr1_-_153363452 | 0.05 |
ENST00000368732.1
ENST00000368733.3 |
S100A8
|
S100 calcium binding protein A8 |
| chr5_+_140625147 | 0.05 |
ENST00000231173.3
|
PCDHB15
|
protocadherin beta 15 |
| chr2_+_87754887 | 0.05 |
ENST00000409054.1
ENST00000331944.6 ENST00000409139.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr14_+_79745682 | 0.04 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr2_-_40657397 | 0.04 |
ENST00000408028.2
ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr9_+_35538616 | 0.04 |
ENST00000455600.1
|
RUSC2
|
RUN and SH3 domain containing 2 |
| chr15_-_54025300 | 0.04 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr6_+_42584847 | 0.03 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr8_+_24151620 | 0.03 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr11_+_9685604 | 0.02 |
ENST00000447399.2
ENST00000318950.6 |
SWAP70
|
SWAP switching B-cell complex 70kDa subunit |
| chr12_-_31743901 | 0.02 |
ENST00000354285.4
|
DENND5B
|
DENN/MADD domain containing 5B |
| chrX_+_55649833 | 0.02 |
ENST00000339140.3
|
FOXR2
|
forkhead box R2 |
| chr18_-_53804580 | 0.02 |
ENST00000590484.1
ENST00000589293.1 ENST00000587904.1 ENST00000591974.1 |
RP11-456O19.4
|
RP11-456O19.4 |
| chr16_+_1877204 | 0.01 |
ENST00000427358.2
|
FAHD1
|
fumarylacetoacetate hydrolase domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HDX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.7 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.7 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |