Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
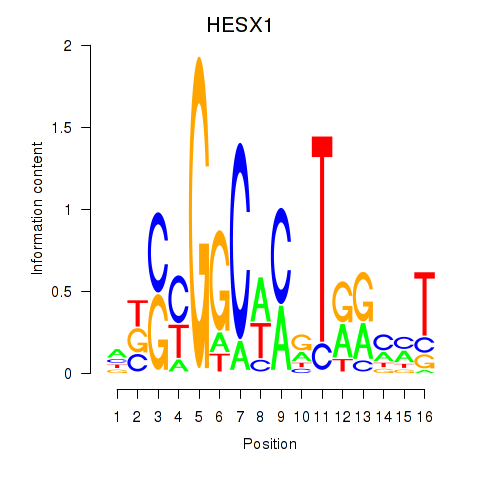
Results for HESX1
Z-value: 0.98
Transcription factors associated with HESX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HESX1
|
ENSG00000163666.4 | HESX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HESX1 | hg19_v2_chr3_-_57233966_57234014 | -0.35 | 5.8e-02 | Click! |
Activity profile of HESX1 motif
Sorted Z-values of HESX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_161337662 | 6.86 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr11_+_101918153 | 5.14 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr6_+_52285131 | 5.01 |
ENST00000433625.2
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr6_+_116937636 | 4.07 |
ENST00000368581.4
ENST00000229554.5 ENST00000368580.4 |
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr1_-_42921915 | 3.86 |
ENST00000372565.3
ENST00000433602.2 |
ZMYND12
|
zinc finger, MYND-type containing 12 |
| chr6_+_52285046 | 3.46 |
ENST00000371068.5
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr16_+_19421803 | 3.41 |
ENST00000541464.1
|
TMC5
|
transmembrane channel-like 5 |
| chr11_+_71791359 | 2.92 |
ENST00000419228.1
ENST00000435085.1 ENST00000307198.7 ENST00000538413.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr16_+_4784273 | 2.92 |
ENST00000299320.5
ENST00000586724.1 |
C16orf71
|
chromosome 16 open reading frame 71 |
| chr8_-_110656995 | 2.87 |
ENST00000276646.9
ENST00000533065.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr8_+_120885949 | 2.74 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr2_-_160654745 | 2.48 |
ENST00000259053.4
ENST00000429078.2 |
CD302
|
CD302 molecule |
| chr1_+_15256230 | 2.41 |
ENST00000376028.4
ENST00000400798.2 |
KAZN
|
kazrin, periplakin interacting protein |
| chr11_-_5276008 | 2.11 |
ENST00000336906.4
|
HBG2
|
hemoglobin, gamma G |
| chr21_+_33784670 | 2.03 |
ENST00000300255.2
|
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr2_+_230787213 | 2.03 |
ENST00000409992.1
|
FBXO36
|
F-box protein 36 |
| chr16_-_84273304 | 2.01 |
ENST00000308251.4
ENST00000568181.1 |
KCNG4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chr1_+_3614591 | 1.98 |
ENST00000378290.4
|
TP73
|
tumor protein p73 |
| chr2_+_230787201 | 1.97 |
ENST00000283946.3
|
FBXO36
|
F-box protein 36 |
| chr5_+_149340282 | 1.90 |
ENST00000286298.4
|
SLC26A2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr2_+_85981008 | 1.88 |
ENST00000306279.3
|
ATOH8
|
atonal homolog 8 (Drosophila) |
| chr7_-_95025661 | 1.83 |
ENST00000542556.1
ENST00000265627.5 ENST00000427422.1 ENST00000451904.1 |
PON1
PON3
|
paraoxonase 1 paraoxonase 3 |
| chr20_-_40247133 | 1.71 |
ENST00000373233.3
ENST00000309279.7 |
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr6_+_116421976 | 1.70 |
ENST00000319550.4
ENST00000419791.1 |
NT5DC1
|
5'-nucleotidase domain containing 1 |
| chr15_-_89089860 | 1.61 |
ENST00000558413.1
ENST00000564406.1 ENST00000268148.8 |
DET1
|
de-etiolated homolog 1 (Arabidopsis) |
| chr2_+_186603355 | 1.53 |
ENST00000343098.5
|
FSIP2
|
fibrous sheath interacting protein 2 |
| chr2_-_43823093 | 1.51 |
ENST00000405006.4
|
THADA
|
thyroid adenoma associated |
| chr9_+_1051481 | 1.46 |
ENST00000358146.2
ENST00000259622.6 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr9_-_123239632 | 1.45 |
ENST00000416449.1
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr16_+_4784458 | 1.44 |
ENST00000590191.1
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr16_-_1661988 | 1.43 |
ENST00000426508.2
|
IFT140
|
intraflagellar transport 140 homolog (Chlamydomonas) |
| chr12_-_49582593 | 1.39 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr19_+_11457232 | 1.39 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr12_+_131438443 | 1.37 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr19_+_45312347 | 1.35 |
ENST00000270233.6
ENST00000591520.1 |
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr20_-_34117447 | 1.34 |
ENST00000246199.2
ENST00000424444.1 ENST00000374345.4 ENST00000444723.1 |
C20orf173
|
chromosome 20 open reading frame 173 |
| chr12_-_121019165 | 1.34 |
ENST00000341039.2
ENST00000357500.4 |
POP5
|
processing of precursor 5, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr19_-_45579762 | 1.22 |
ENST00000303809.2
|
ZNF296
|
zinc finger protein 296 |
| chr21_-_34852304 | 1.19 |
ENST00000542230.2
|
TMEM50B
|
transmembrane protein 50B |
| chr1_-_169337176 | 1.15 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr1_+_156338993 | 1.13 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr17_-_57184260 | 1.08 |
ENST00000376149.3
ENST00000393066.3 |
TRIM37
|
tripartite motif containing 37 |
| chr6_-_41888814 | 1.05 |
ENST00000409060.1
ENST00000265350.4 |
MED20
|
mediator complex subunit 20 |
| chr7_-_99679324 | 1.03 |
ENST00000292393.5
ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3
|
zinc finger protein 3 |
| chr8_-_97273807 | 0.98 |
ENST00000517720.1
ENST00000287025.3 ENST00000523821.1 |
MTERFD1
|
MTERF domain containing 1 |
| chr12_-_55367361 | 0.96 |
ENST00000532804.1
ENST00000531122.1 ENST00000533446.1 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr5_-_122758994 | 0.96 |
ENST00000306467.5
ENST00000515110.1 |
CEP120
|
centrosomal protein 120kDa |
| chr9_+_37079888 | 0.96 |
ENST00000429493.1
ENST00000593237.1 ENST00000588557.1 ENST00000430809.1 ENST00000592157.1 |
RP11-220I1.1
|
RP11-220I1.1 |
| chr7_+_74508372 | 0.91 |
ENST00000356115.5
ENST00000430511.2 ENST00000312575.7 |
GTF2IRD2B
|
GTF2I repeat domain containing 2B |
| chr11_+_73019282 | 0.91 |
ENST00000263674.3
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr14_-_70883708 | 0.88 |
ENST00000256366.4
|
SYNJ2BP
|
synaptojanin 2 binding protein |
| chr11_-_1629693 | 0.88 |
ENST00000399685.1
|
KRTAP5-3
|
keratin associated protein 5-3 |
| chr4_+_6784401 | 0.88 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chr3_-_196695692 | 0.86 |
ENST00000412723.1
|
PIGZ
|
phosphatidylinositol glycan anchor biosynthesis, class Z |
| chr17_+_55163075 | 0.83 |
ENST00000571629.1
ENST00000570423.1 ENST00000575186.1 ENST00000573085.1 ENST00000572814.1 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr13_-_52378231 | 0.83 |
ENST00000280056.2
ENST00000444610.2 |
DHRS12
|
dehydrogenase/reductase (SDR family) member 12 |
| chr2_-_43823119 | 0.82 |
ENST00000403856.1
ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA
|
thyroid adenoma associated |
| chr1_-_109968973 | 0.81 |
ENST00000271308.4
ENST00000538610.1 |
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5 |
| chr10_+_90672113 | 0.76 |
ENST00000371922.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr3_-_51975942 | 0.76 |
ENST00000232888.6
|
RRP9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr22_+_18632666 | 0.75 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr2_+_233562015 | 0.75 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr6_-_41888843 | 0.74 |
ENST00000434077.1
ENST00000409312.1 |
MED20
|
mediator complex subunit 20 |
| chr16_-_70719925 | 0.74 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr11_+_126139005 | 0.74 |
ENST00000263578.5
ENST00000442061.2 ENST00000532125.1 |
FOXRED1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr4_-_2264015 | 0.71 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr6_-_83775489 | 0.69 |
ENST00000369747.3
|
UBE3D
|
ubiquitin protein ligase E3D |
| chrX_-_7895479 | 0.69 |
ENST00000381042.4
|
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr9_-_34376851 | 0.69 |
ENST00000297625.7
|
KIAA1161
|
KIAA1161 |
| chr3_+_195943369 | 0.68 |
ENST00000296327.5
|
SLC51A
|
solute carrier family 51, alpha subunit |
| chr6_-_167275991 | 0.68 |
ENST00000510118.1
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr10_+_103348031 | 0.68 |
ENST00000370151.4
ENST00000370147.1 ENST00000370148.2 |
DPCD
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr17_+_36861735 | 0.67 |
ENST00000378137.5
ENST00000325718.7 |
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr3_+_192958914 | 0.66 |
ENST00000264735.2
ENST00000602513.1 |
HRASLS
|
HRAS-like suppressor |
| chr5_-_122759032 | 0.65 |
ENST00000510582.3
ENST00000328236.5 ENST00000306481.6 ENST00000508442.2 ENST00000395431.2 |
CEP120
|
centrosomal protein 120kDa |
| chr7_-_74267836 | 0.59 |
ENST00000361071.5
ENST00000453619.2 ENST00000417115.2 ENST00000405086.2 |
GTF2IRD2
|
GTF2I repeat domain containing 2 |
| chr2_-_89340242 | 0.57 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr21_+_46020497 | 0.55 |
ENST00000380102.2
|
KRTAP10-7
|
keratin associated protein 10-7 |
| chr2_+_220071490 | 0.54 |
ENST00000409206.1
ENST00000409594.1 ENST00000289528.5 ENST00000422255.1 ENST00000409412.1 ENST00000409097.1 ENST00000409336.1 ENST00000409217.1 ENST00000409319.1 ENST00000444522.2 |
ZFAND2B
|
zinc finger, AN1-type domain 2B |
| chr2_-_89513402 | 0.54 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr14_-_25103388 | 0.53 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr16_-_57219966 | 0.52 |
ENST00000565760.1
ENST00000309137.8 ENST00000570184.1 ENST00000562324.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr14_-_90798273 | 0.51 |
ENST00000357904.3
|
NRDE2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr4_+_129349188 | 0.50 |
ENST00000511497.1
|
RP11-420A23.1
|
RP11-420A23.1 |
| chr13_+_108870714 | 0.49 |
ENST00000375898.3
|
ABHD13
|
abhydrolase domain containing 13 |
| chr2_-_89399845 | 0.49 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr4_-_89619386 | 0.48 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr19_+_46498704 | 0.48 |
ENST00000595358.1
ENST00000594672.1 ENST00000536603.1 |
CCDC61
|
coiled-coil domain containing 61 |
| chr16_+_66914264 | 0.45 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr17_-_57184064 | 0.45 |
ENST00000262294.7
|
TRIM37
|
tripartite motif containing 37 |
| chr4_+_175204818 | 0.44 |
ENST00000503780.1
|
CEP44
|
centrosomal protein 44kDa |
| chr13_+_100741269 | 0.44 |
ENST00000376286.4
ENST00000376279.3 ENST00000376285.1 |
PCCA
|
propionyl CoA carboxylase, alpha polypeptide |
| chr11_-_4414880 | 0.43 |
ENST00000254436.7
ENST00000543625.1 |
TRIM21
|
tripartite motif containing 21 |
| chr19_+_38397839 | 0.43 |
ENST00000222345.6
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3 |
| chr2_+_90198535 | 0.42 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr8_-_145060593 | 0.42 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr16_-_57219721 | 0.42 |
ENST00000562406.1
ENST00000568671.1 ENST00000567044.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr14_-_25103472 | 0.41 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr3_-_131753830 | 0.41 |
ENST00000429747.1
|
CPNE4
|
copine IV |
| chr12_-_123717643 | 0.41 |
ENST00000541437.1
ENST00000606320.1 |
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr1_+_152658599 | 0.40 |
ENST00000368780.3
|
LCE2B
|
late cornified envelope 2B |
| chr7_-_37024665 | 0.39 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr16_-_57219926 | 0.39 |
ENST00000566584.1
ENST00000566481.1 ENST00000566077.1 ENST00000564108.1 ENST00000565458.1 ENST00000566681.1 ENST00000567439.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr8_+_124084899 | 0.38 |
ENST00000287380.1
ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31
|
TBC1 domain family, member 31 |
| chr8_+_146277764 | 0.37 |
ENST00000331434.6
|
C8orf33
|
chromosome 8 open reading frame 33 |
| chr17_+_18128896 | 0.37 |
ENST00000316843.4
|
LLGL1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr3_+_155860751 | 0.37 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chrX_-_64754611 | 0.37 |
ENST00000374807.5
ENST00000374811.3 ENST00000374804.5 ENST00000312391.8 |
LAS1L
|
LAS1-like (S. cerevisiae) |
| chr19_-_39694894 | 0.37 |
ENST00000318438.6
|
SYCN
|
syncollin |
| chr5_-_176057518 | 0.36 |
ENST00000393693.2
|
SNCB
|
synuclein, beta |
| chr21_+_38445539 | 0.35 |
ENST00000418766.1
ENST00000450533.1 ENST00000438055.1 ENST00000355666.1 ENST00000540756.1 ENST00000399010.1 |
TTC3
|
tetratricopeptide repeat domain 3 |
| chr20_-_61002584 | 0.34 |
ENST00000252998.1
|
RBBP8NL
|
RBBP8 N-terminal like |
| chr17_+_46018872 | 0.33 |
ENST00000583599.1
ENST00000434554.2 ENST00000225573.4 ENST00000544840.1 ENST00000534893.1 |
PNPO
|
pyridoxamine 5'-phosphate oxidase |
| chr7_+_1022811 | 0.31 |
ENST00000308919.7
|
CYP2W1
|
cytochrome P450, family 2, subfamily W, polypeptide 1 |
| chr11_-_126138808 | 0.29 |
ENST00000332118.6
ENST00000532259.1 |
SRPR
|
signal recognition particle receptor (docking protein) |
| chr8_+_71581565 | 0.28 |
ENST00000408926.3
ENST00000520030.1 |
XKR9
|
XK, Kell blood group complex subunit-related family, member 9 |
| chr16_+_1662326 | 0.28 |
ENST00000397412.3
|
CRAMP1L
|
Crm, cramped-like (Drosophila) |
| chr2_+_90192768 | 0.27 |
ENST00000390275.2
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr8_+_109455845 | 0.26 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr20_-_5485166 | 0.26 |
ENST00000589201.1
ENST00000379053.4 |
LINC00654
|
long intergenic non-protein coding RNA 654 |
| chr2_-_96931679 | 0.26 |
ENST00000258439.3
ENST00000432959.1 |
TMEM127
|
transmembrane protein 127 |
| chr14_-_90798418 | 0.25 |
ENST00000354366.3
|
NRDE2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr19_+_41699135 | 0.25 |
ENST00000542619.1
ENST00000600561.1 |
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr15_-_83316711 | 0.24 |
ENST00000568128.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr12_-_53228079 | 0.23 |
ENST00000330553.5
|
KRT79
|
keratin 79 |
| chr1_-_165738072 | 0.23 |
ENST00000481278.1
|
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr4_-_175204765 | 0.23 |
ENST00000513696.1
ENST00000503293.1 |
FBXO8
|
F-box protein 8 |
| chr13_-_108870623 | 0.23 |
ENST00000405925.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr18_+_54318566 | 0.23 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr2_-_128051670 | 0.22 |
ENST00000493187.2
|
ERCC3
|
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
| chr6_+_56820018 | 0.22 |
ENST00000370746.3
|
BEND6
|
BEN domain containing 6 |
| chr5_+_78532003 | 0.21 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr11_+_47586982 | 0.21 |
ENST00000426530.2
ENST00000534775.1 |
PTPMT1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr20_+_47835884 | 0.21 |
ENST00000371764.4
|
DDX27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr4_+_15683404 | 0.21 |
ENST00000422728.2
|
FAM200B
|
family with sequence similarity 200, member B |
| chr16_-_14724057 | 0.20 |
ENST00000539279.1
ENST00000420015.2 ENST00000437198.2 |
PARN
|
poly(A)-specific ribonuclease |
| chr21_-_38445470 | 0.20 |
ENST00000399098.1
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr1_-_54637997 | 0.20 |
ENST00000536061.1
|
AL357673.1
|
CDNA: FLJ21031 fis, clone CAE07336; HCG1780521; Uncharacterized protein |
| chr21_+_34398153 | 0.19 |
ENST00000382357.3
ENST00000430860.1 ENST00000333337.3 |
OLIG2
|
oligodendrocyte lineage transcription factor 2 |
| chr4_+_15683369 | 0.19 |
ENST00000503617.1
|
FAM200B
|
family with sequence similarity 200, member B |
| chr6_-_42981651 | 0.19 |
ENST00000244711.3
|
MEA1
|
male-enhanced antigen 1 |
| chr15_+_32322709 | 0.18 |
ENST00000455693.2
|
CHRNA7
|
cholinergic receptor, nicotinic, alpha 7 (neuronal) |
| chr10_+_121652204 | 0.18 |
ENST00000369075.3
ENST00000543134.1 |
SEC23IP
|
SEC23 interacting protein |
| chr7_+_1022973 | 0.18 |
ENST00000340150.6
|
CYP2W1
|
cytochrome P450, family 2, subfamily W, polypeptide 1 |
| chr2_+_32502952 | 0.17 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr21_-_38445011 | 0.17 |
ENST00000464265.1
ENST00000399102.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr11_+_67222818 | 0.17 |
ENST00000325656.5
|
CABP4
|
calcium binding protein 4 |
| chr1_-_20446020 | 0.17 |
ENST00000375105.3
|
PLA2G2D
|
phospholipase A2, group IID |
| chr19_+_19431490 | 0.16 |
ENST00000392313.6
ENST00000262815.8 ENST00000609122.1 |
MAU2
|
MAU2 sister chromatid cohesion factor |
| chrX_+_118602363 | 0.16 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr6_+_56819773 | 0.16 |
ENST00000370750.2
|
BEND6
|
BEN domain containing 6 |
| chr18_+_56892724 | 0.15 |
ENST00000456142.3
ENST00000530323.1 |
GRP
|
gastrin-releasing peptide |
| chr3_+_178253993 | 0.15 |
ENST00000420517.2
ENST00000452583.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr10_+_26505594 | 0.15 |
ENST00000259271.3
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr17_-_15168624 | 0.14 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr1_-_165738085 | 0.14 |
ENST00000464650.1
ENST00000392129.6 |
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr3_+_138153451 | 0.14 |
ENST00000389567.4
ENST00000289135.4 |
ESYT3
|
extended synaptotagmin-like protein 3 |
| chrX_+_135730373 | 0.14 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr8_-_101734308 | 0.13 |
ENST00000519004.1
ENST00000519363.1 ENST00000520142.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr11_+_47587129 | 0.13 |
ENST00000326656.8
ENST00000326674.9 |
PTPMT1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr2_+_3705785 | 0.12 |
ENST00000252505.3
|
ALLC
|
allantoicase |
| chr14_+_45553296 | 0.12 |
ENST00000355765.6
ENST00000553605.1 |
PRPF39
|
pre-mRNA processing factor 39 |
| chr7_+_99425633 | 0.12 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chrX_+_135730297 | 0.12 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr1_-_16302565 | 0.11 |
ENST00000537142.1
ENST00000448462.2 |
ZBTB17
|
zinc finger and BTB domain containing 17 |
| chr2_-_197036289 | 0.11 |
ENST00000263955.4
|
STK17B
|
serine/threonine kinase 17b |
| chr1_-_1356628 | 0.10 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr12_-_54070098 | 0.10 |
ENST00000394349.3
ENST00000549164.1 |
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr12_-_121476750 | 0.09 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr8_-_101734170 | 0.09 |
ENST00000522387.1
ENST00000518196.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_+_47799446 | 0.09 |
ENST00000371873.5
|
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr2_+_234216454 | 0.09 |
ENST00000447536.1
ENST00000409110.1 |
SAG
|
S-antigen; retina and pineal gland (arrestin) |
| chr19_+_34919257 | 0.09 |
ENST00000246548.4
ENST00000590048.2 |
UBA2
|
ubiquitin-like modifier activating enzyme 2 |
| chr1_+_46016703 | 0.09 |
ENST00000481885.1
ENST00000351829.4 ENST00000471651.1 |
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr16_+_56995762 | 0.08 |
ENST00000200676.3
ENST00000379780.2 |
CETP
|
cholesteryl ester transfer protein, plasma |
| chr18_-_44702668 | 0.08 |
ENST00000256433.3
|
IER3IP1
|
immediate early response 3 interacting protein 1 |
| chr7_+_23637118 | 0.07 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr2_-_74875432 | 0.06 |
ENST00000536235.1
ENST00000421985.1 |
M1AP
|
meiosis 1 associated protein |
| chr2_-_61765315 | 0.06 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr15_+_22892663 | 0.06 |
ENST00000313077.7
ENST00000561274.1 ENST00000560848.1 |
CYFIP1
|
cytoplasmic FMR1 interacting protein 1 |
| chr20_+_35807512 | 0.05 |
ENST00000373622.5
|
RPN2
|
ribophorin II |
| chr6_-_29600832 | 0.05 |
ENST00000377016.4
ENST00000376977.3 ENST00000377034.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr4_-_48782259 | 0.05 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr16_-_20367584 | 0.04 |
ENST00000570689.1
|
UMOD
|
uromodulin |
| chr7_+_99746514 | 0.04 |
ENST00000341942.5
ENST00000441173.1 |
LAMTOR4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr20_-_62587735 | 0.03 |
ENST00000354216.6
ENST00000369892.3 ENST00000358711.3 |
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr6_-_110797799 | 0.03 |
ENST00000456137.2
ENST00000368919.3 ENST00000439654.1 |
SLC22A16
|
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
| chr17_+_45286387 | 0.03 |
ENST00000572316.1
ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr21_-_38445443 | 0.02 |
ENST00000360525.4
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr22_+_31892373 | 0.02 |
ENST00000443011.1
ENST00000400289.1 ENST00000444859.1 ENST00000400288.2 |
SFI1
|
Sfi1 homolog, spindle assembly associated (yeast) |
| chr22_+_19419425 | 0.02 |
ENST00000333130.3
|
MRPL40
|
mitochondrial ribosomal protein L40 |
| chr4_+_83956237 | 0.01 |
ENST00000264389.2
|
COPS4
|
COP9 signalosome subunit 4 |
| chr11_+_118443098 | 0.01 |
ENST00000392859.3
ENST00000359415.4 ENST00000534182.2 ENST00000264028.4 |
ARCN1
|
archain 1 |
| chr15_-_51058005 | 0.00 |
ENST00000261854.5
|
SPPL2A
|
signal peptide peptidase like 2A |
| chr20_+_35807449 | 0.00 |
ENST00000237530.6
|
RPN2
|
ribophorin II |
| chr5_-_159546396 | 0.00 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chrX_+_153409678 | 0.00 |
ENST00000369951.4
|
OPN1LW
|
opsin 1 (cone pigments), long-wave-sensitive |
Network of associatons between targets according to the STRING database.
First level regulatory network of HESX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.5 | 1.5 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.5 | 1.8 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) phenylpropanoid catabolic process(GO:0046271) |
| 0.4 | 1.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.2 | 0.5 | GO:0046222 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.2 | 1.5 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 1.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.2 | 1.9 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 1.6 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 1.7 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 2.9 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 2.7 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 1.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.9 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 1.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 1.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 8.5 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.1 | 0.4 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 | 0.4 | GO:0010847 | regulation of chromatin assembly(GO:0010847) negative regulation of protein import into nucleus, translocation(GO:0033159) protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 2.0 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.1 | 2.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.9 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.9 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.7 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.1 | 0.3 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.5 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.2 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 | 0.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 4.1 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.2 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 1.3 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.4 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.2 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.7 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0043605 | allantoin metabolic process(GO:0000255) cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 1.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.8 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.2 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 2.5 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.4 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.0 | 0.4 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 2.6 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.7 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.2 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 1.0 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.7 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.7 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.7 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.3 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.2 | 2.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.2 | 2.9 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 1.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 1.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 0.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 2.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 1.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.8 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 8.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 2.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 1.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.3 | 2.9 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.3 | 0.9 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.3 | 1.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.2 | 1.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 2.0 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.7 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.8 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 2.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.3 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 1.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 2.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 2.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.2 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 1.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.1 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 1.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 9.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.8 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.4 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.5 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.5 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 4.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 2.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 2.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 2.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 3.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |