Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
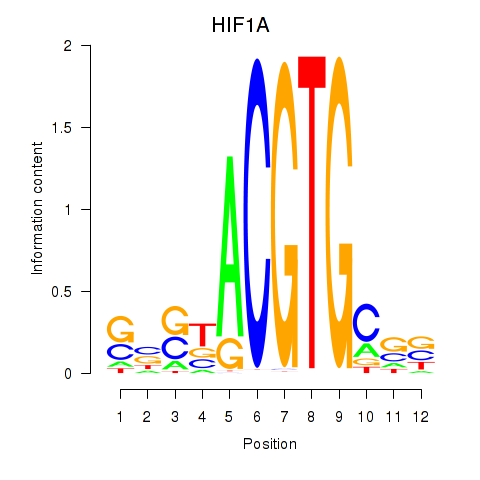
Results for HIF1A
Z-value: 0.82
Transcription factors associated with HIF1A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIF1A
|
ENSG00000100644.12 | hypoxia inducible factor 1 subunit alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIF1A | hg19_v2_chr14_+_62162258_62162269 | -0.45 | 1.2e-02 | Click! |
Activity profile of HIF1A motif
Sorted Z-values of HIF1A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_19988462 | 3.69 |
ENST00000344838.4
|
EFHB
|
EF-hand domain family, member B |
| chr5_+_10441970 | 2.10 |
ENST00000274134.4
|
ROPN1L
|
rhophilin associated tail protein 1-like |
| chr4_-_186317034 | 1.97 |
ENST00000505916.1
|
LRP2BP
|
LRP2 binding protein |
| chr1_+_150254936 | 1.89 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr8_+_75896731 | 1.73 |
ENST00000262207.4
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1 |
| chr7_+_48075108 | 1.67 |
ENST00000420324.1
ENST00000435376.1 ENST00000430738.1 ENST00000348904.3 ENST00000539619.1 |
C7orf57
|
chromosome 7 open reading frame 57 |
| chr12_+_7023735 | 1.62 |
ENST00000538763.1
ENST00000544774.1 ENST00000545045.2 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr12_+_7023491 | 1.61 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr2_-_28113965 | 1.54 |
ENST00000302188.3
|
RBKS
|
ribokinase |
| chr2_-_220083076 | 1.25 |
ENST00000295750.4
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr2_-_28113217 | 1.24 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr11_+_74204395 | 1.19 |
ENST00000526036.1
|
AP001372.2
|
AP001372.2 |
| chr3_-_145878954 | 1.18 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr19_+_5720666 | 1.15 |
ENST00000381624.3
ENST00000381614.2 |
CATSPERD
|
catsper channel auxiliary subunit delta |
| chr8_+_17354617 | 1.11 |
ENST00000470360.1
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr10_+_120789223 | 1.08 |
ENST00000425699.1
|
NANOS1
|
nanos homolog 1 (Drosophila) |
| chr19_-_55691614 | 1.06 |
ENST00000592470.1
ENST00000354308.3 |
SYT5
|
synaptotagmin V |
| chrX_+_69509927 | 0.99 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chr8_+_17354587 | 0.96 |
ENST00000494857.1
ENST00000522656.1 |
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr14_-_64970494 | 0.93 |
ENST00000608382.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr1_+_214454492 | 0.90 |
ENST00000366957.5
ENST00000415093.2 |
SMYD2
|
SET and MYND domain containing 2 |
| chr10_-_93392811 | 0.87 |
ENST00000238994.5
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C |
| chr2_+_173420697 | 0.85 |
ENST00000282077.3
ENST00000392571.2 ENST00000410055.1 |
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr2_-_216946500 | 0.84 |
ENST00000265322.7
|
PECR
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr4_-_39529049 | 0.80 |
ENST00000501493.2
ENST00000509391.1 ENST00000507089.1 |
UGDH
|
UDP-glucose 6-dehydrogenase |
| chr11_-_76155700 | 0.79 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr17_-_34890759 | 0.78 |
ENST00000431794.3
|
MYO19
|
myosin XIX |
| chr5_-_146889619 | 0.77 |
ENST00000343218.5
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr4_-_39529180 | 0.77 |
ENST00000515021.1
ENST00000510490.1 ENST00000316423.6 |
UGDH
|
UDP-glucose 6-dehydrogenase |
| chr12_-_49318715 | 0.76 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr2_-_68384603 | 0.75 |
ENST00000406245.2
ENST00000409164.1 ENST00000295121.6 |
WDR92
|
WD repeat domain 92 |
| chr3_+_113666748 | 0.74 |
ENST00000330212.3
ENST00000498275.1 |
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr11_+_7534999 | 0.72 |
ENST00000528947.1
ENST00000299492.4 |
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr4_+_129732467 | 0.71 |
ENST00000413543.2
|
PHF17
|
jade family PHD finger 1 |
| chr22_+_41777927 | 0.67 |
ENST00000266304.4
|
TEF
|
thyrotrophic embryonic factor |
| chr2_-_220083671 | 0.66 |
ENST00000439002.2
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr18_+_43913919 | 0.65 |
ENST00000587853.1
|
RNF165
|
ring finger protein 165 |
| chr2_-_220083692 | 0.65 |
ENST00000265316.3
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr22_-_24181174 | 0.64 |
ENST00000318109.7
ENST00000406855.3 ENST00000404056.1 ENST00000476077.1 |
DERL3
|
derlin 3 |
| chr11_-_76155618 | 0.63 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chrX_-_13956737 | 0.63 |
ENST00000454189.2
|
GPM6B
|
glycoprotein M6B |
| chr3_+_100211412 | 0.61 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr8_+_26240414 | 0.60 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr13_-_24463530 | 0.58 |
ENST00000382172.3
|
MIPEP
|
mitochondrial intermediate peptidase |
| chr6_+_35265586 | 0.56 |
ENST00000542066.1
ENST00000316637.5 |
DEF6
|
differentially expressed in FDCP 6 homolog (mouse) |
| chrX_-_34675391 | 0.56 |
ENST00000275954.3
|
TMEM47
|
transmembrane protein 47 |
| chr11_-_118901559 | 0.55 |
ENST00000330775.7
ENST00000545985.1 ENST00000357590.5 ENST00000538950.1 |
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr1_+_100436065 | 0.55 |
ENST00000370153.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr7_+_128095945 | 0.54 |
ENST00000257696.4
|
HILPDA
|
hypoxia inducible lipid droplet-associated |
| chr6_-_83775489 | 0.54 |
ENST00000369747.3
|
UBE3D
|
ubiquitin protein ligase E3D |
| chr16_-_30022735 | 0.54 |
ENST00000564944.1
|
DOC2A
|
double C2-like domains, alpha |
| chr2_+_27665232 | 0.53 |
ENST00000543753.1
ENST00000288873.3 |
KRTCAP3
|
keratinocyte associated protein 3 |
| chr12_+_110906169 | 0.52 |
ENST00000377673.5
|
FAM216A
|
family with sequence similarity 216, member A |
| chr15_-_60884706 | 0.49 |
ENST00000449337.2
|
RORA
|
RAR-related orphan receptor A |
| chr20_+_30946106 | 0.49 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr4_-_104119528 | 0.49 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr1_-_231376836 | 0.49 |
ENST00000451322.1
|
C1orf131
|
chromosome 1 open reading frame 131 |
| chr19_-_1568057 | 0.48 |
ENST00000402693.4
ENST00000388824.6 |
MEX3D
|
mex-3 RNA binding family member D |
| chr17_+_7608511 | 0.48 |
ENST00000226091.2
|
EFNB3
|
ephrin-B3 |
| chr2_+_27665289 | 0.48 |
ENST00000407293.1
|
KRTCAP3
|
keratinocyte associated protein 3 |
| chr14_+_64970662 | 0.48 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chrX_-_13956497 | 0.46 |
ENST00000398361.3
|
GPM6B
|
glycoprotein M6B |
| chr11_-_76381781 | 0.46 |
ENST00000260061.5
ENST00000404995.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chrX_+_77359671 | 0.45 |
ENST00000373316.4
|
PGK1
|
phosphoglycerate kinase 1 |
| chr9_-_139922726 | 0.45 |
ENST00000265662.5
ENST00000371605.3 |
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr15_+_41056218 | 0.45 |
ENST00000260447.4
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr2_+_10183651 | 0.44 |
ENST00000305883.1
|
KLF11
|
Kruppel-like factor 11 |
| chr7_+_116593433 | 0.44 |
ENST00000323984.3
ENST00000393449.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr19_-_49137790 | 0.44 |
ENST00000599385.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr19_-_49137762 | 0.43 |
ENST00000593500.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr6_+_151561085 | 0.43 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr15_+_41056255 | 0.43 |
ENST00000561160.1
ENST00000559445.1 |
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr1_-_92351769 | 0.42 |
ENST00000212355.4
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr1_-_159894319 | 0.42 |
ENST00000320307.4
|
TAGLN2
|
transgelin 2 |
| chr11_-_17498348 | 0.42 |
ENST00000389817.3
ENST00000302539.4 |
ABCC8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
| chr16_-_30022293 | 0.41 |
ENST00000565273.1
ENST00000567332.2 ENST00000350119.4 |
DOC2A
|
double C2-like domains, alpha |
| chr2_-_97652324 | 0.41 |
ENST00000490605.2
|
FAM178B
|
family with sequence similarity 178, member B |
| chrX_+_77359726 | 0.41 |
ENST00000442431.1
|
PGK1
|
phosphoglycerate kinase 1 |
| chr7_+_26331541 | 0.40 |
ENST00000416246.1
ENST00000338523.4 ENST00000412416.1 |
SNX10
|
sorting nexin 10 |
| chr3_+_49044798 | 0.40 |
ENST00000438660.1
ENST00000608424.1 ENST00000415265.2 |
WDR6
|
WD repeat domain 6 |
| chr6_-_153304148 | 0.40 |
ENST00000229758.3
|
FBXO5
|
F-box protein 5 |
| chr7_+_128095900 | 0.40 |
ENST00000435296.2
|
HILPDA
|
hypoxia inducible lipid droplet-associated |
| chr17_-_57184260 | 0.39 |
ENST00000376149.3
ENST00000393066.3 |
TRIM37
|
tripartite motif containing 37 |
| chr11_+_9595180 | 0.39 |
ENST00000450114.2
|
WEE1
|
WEE1 G2 checkpoint kinase |
| chr2_-_97652290 | 0.39 |
ENST00000327896.3
|
FAM178B
|
family with sequence similarity 178, member B |
| chr16_+_66878814 | 0.39 |
ENST00000394069.3
|
CA7
|
carbonic anhydrase VII |
| chr8_-_103425047 | 0.38 |
ENST00000520539.1
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr2_+_216946589 | 0.38 |
ENST00000433112.1
ENST00000454545.1 ENST00000437356.2 ENST00000295658.4 ENST00000455479.1 ENST00000406027.2 |
TMEM169
|
transmembrane protein 169 |
| chr1_-_26233423 | 0.37 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr16_-_54963026 | 0.37 |
ENST00000560208.1
ENST00000557792.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr6_+_87865262 | 0.37 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr19_+_41903709 | 0.37 |
ENST00000542943.1
ENST00000457836.2 |
BCKDHA
|
branched chain keto acid dehydrogenase E1, alpha polypeptide |
| chr14_-_24685246 | 0.36 |
ENST00000396833.2
ENST00000288087.7 |
MDP1
|
magnesium-dependent phosphatase 1 |
| chr1_+_22333943 | 0.36 |
ENST00000400271.2
|
CELA3A
|
chymotrypsin-like elastase family, member 3A |
| chr17_+_80416050 | 0.36 |
ENST00000579198.1
ENST00000390006.4 ENST00000580296.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr17_-_73127353 | 0.36 |
ENST00000580423.1
ENST00000578337.1 ENST00000582160.1 |
NT5C
|
5', 3'-nucleotidase, cytosolic |
| chr11_+_73498898 | 0.36 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr12_-_121734489 | 0.36 |
ENST00000412367.2
ENST00000402834.4 ENST00000404169.3 |
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr17_-_73127826 | 0.36 |
ENST00000582170.1
ENST00000245552.2 |
NT5C
|
5', 3'-nucleotidase, cytosolic |
| chr3_+_25831567 | 0.35 |
ENST00000280701.3
ENST00000420173.2 |
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr13_+_27998681 | 0.35 |
ENST00000381140.4
|
GTF3A
|
general transcription factor IIIA |
| chr17_+_76311791 | 0.35 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr11_-_63933504 | 0.34 |
ENST00000255681.6
|
MACROD1
|
MACRO domain containing 1 |
| chr8_-_95908902 | 0.34 |
ENST00000520509.1
|
CCNE2
|
cyclin E2 |
| chr11_+_6625046 | 0.33 |
ENST00000396751.2
|
ILK
|
integrin-linked kinase |
| chr10_+_49514698 | 0.33 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr9_-_139922631 | 0.33 |
ENST00000341511.6
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr11_+_6624955 | 0.33 |
ENST00000299421.4
ENST00000537806.1 |
ILK
|
integrin-linked kinase |
| chr2_+_88991162 | 0.33 |
ENST00000283646.4
|
RPIA
|
ribose 5-phosphate isomerase A |
| chr2_+_32288657 | 0.32 |
ENST00000345662.1
|
SPAST
|
spastin |
| chr5_+_126112794 | 0.32 |
ENST00000261366.5
ENST00000395354.1 |
LMNB1
|
lamin B1 |
| chr13_+_103451399 | 0.32 |
ENST00000257336.1
ENST00000448849.2 |
BIVM
|
basic, immunoglobulin-like variable motif containing |
| chr3_-_71834318 | 0.32 |
ENST00000353065.3
|
PROK2
|
prokineticin 2 |
| chr3_+_63898275 | 0.31 |
ENST00000538065.1
|
ATXN7
|
ataxin 7 |
| chr11_+_6624970 | 0.31 |
ENST00000420936.2
ENST00000528995.1 |
ILK
|
integrin-linked kinase |
| chr2_+_32288725 | 0.31 |
ENST00000315285.3
|
SPAST
|
spastin |
| chr4_+_17578815 | 0.31 |
ENST00000226299.4
|
LAP3
|
leucine aminopeptidase 3 |
| chr11_-_18343669 | 0.30 |
ENST00000396253.3
ENST00000349215.3 ENST00000438420.2 |
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr13_+_115079949 | 0.30 |
ENST00000361283.1
|
CHAMP1
|
chromosome alignment maintaining phosphoprotein 1 |
| chr17_+_6918354 | 0.30 |
ENST00000552775.1
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr4_-_54930790 | 0.29 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr6_+_89791507 | 0.28 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr6_-_159240415 | 0.28 |
ENST00000367075.3
|
EZR
|
ezrin |
| chr5_-_131563501 | 0.28 |
ENST00000401867.1
ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr3_-_81811312 | 0.27 |
ENST00000429644.2
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr17_-_67323305 | 0.27 |
ENST00000392677.2
ENST00000593153.1 |
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr14_-_23770683 | 0.27 |
ENST00000561437.1
ENST00000559942.1 ENST00000560913.1 ENST00000559314.1 ENST00000558058.1 |
PPP1R3E
|
protein phosphatase 1, regulatory subunit 3E |
| chr17_-_67323232 | 0.27 |
ENST00000592568.1
ENST00000392676.3 |
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr12_+_28343365 | 0.27 |
ENST00000545336.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr22_-_31742218 | 0.27 |
ENST00000266269.5
ENST00000405309.3 ENST00000351933.4 |
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr19_-_5719860 | 0.27 |
ENST00000590729.1
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr2_+_26915584 | 0.27 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3 |
| chr4_-_17783135 | 0.26 |
ENST00000265018.3
|
FAM184B
|
family with sequence similarity 184, member B |
| chr4_+_37245799 | 0.26 |
ENST00000309447.5
|
KIAA1239
|
KIAA1239 |
| chr9_+_115983808 | 0.26 |
ENST00000374210.6
ENST00000374212.4 |
SLC31A1
|
solute carrier family 31 (copper transporter), member 1 |
| chr17_-_35969409 | 0.26 |
ENST00000394378.2
ENST00000585472.1 ENST00000591288.1 ENST00000502449.2 ENST00000345615.4 ENST00000346661.4 ENST00000585689.1 ENST00000339208.6 |
SYNRG
|
synergin, gamma |
| chr1_+_28052518 | 0.26 |
ENST00000530324.1
ENST00000234549.7 ENST00000373949.1 ENST00000010299.6 |
FAM76A
|
family with sequence similarity 76, member A |
| chr22_+_19701985 | 0.26 |
ENST00000455784.2
ENST00000406395.1 |
SEPT5
|
septin 5 |
| chr3_-_71632894 | 0.26 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr12_+_54332535 | 0.25 |
ENST00000243056.3
|
HOXC13
|
homeobox C13 |
| chr11_-_62414070 | 0.25 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr20_+_33464238 | 0.25 |
ENST00000360596.2
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr7_+_96747030 | 0.25 |
ENST00000360382.4
|
ACN9
|
ACN9 homolog (S. cerevisiae) |
| chr17_+_80416482 | 0.25 |
ENST00000309794.11
ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr2_+_204192942 | 0.25 |
ENST00000295851.5
ENST00000261017.5 |
ABI2
|
abl-interactor 2 |
| chr6_-_153304697 | 0.25 |
ENST00000367241.3
|
FBXO5
|
F-box protein 5 |
| chr15_+_38544476 | 0.25 |
ENST00000299084.4
|
SPRED1
|
sprouty-related, EVH1 domain containing 1 |
| chr9_+_117373486 | 0.25 |
ENST00000288502.4
ENST00000374049.4 |
C9orf91
|
chromosome 9 open reading frame 91 |
| chrX_+_24483338 | 0.24 |
ENST00000379162.4
ENST00000441463.2 |
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3 |
| chr7_+_43152191 | 0.24 |
ENST00000395891.2
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr1_-_109506036 | 0.24 |
ENST00000369976.1
ENST00000356970.2 ENST00000369971.2 ENST00000415331.1 ENST00000357393.4 |
CLCC1
AKNAD1
|
chloride channel CLIC-like 1 AKNA domain containing 1 |
| chr5_+_49962772 | 0.24 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr7_-_100026280 | 0.24 |
ENST00000360951.4
ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1
|
zinc finger, CW type with PWWP domain 1 |
| chr15_-_59665062 | 0.24 |
ENST00000288235.4
|
MYO1E
|
myosin IE |
| chr12_+_28343353 | 0.24 |
ENST00000539107.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr12_-_2113583 | 0.23 |
ENST00000397173.4
ENST00000280665.6 |
DCP1B
|
decapping mRNA 1B |
| chr20_+_33464407 | 0.23 |
ENST00000253382.5
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr16_+_66914264 | 0.23 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr17_+_57642886 | 0.23 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr14_-_20923195 | 0.23 |
ENST00000206542.4
|
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr17_+_72428218 | 0.23 |
ENST00000392628.2
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr7_-_139876812 | 0.22 |
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr21_+_45719921 | 0.22 |
ENST00000349048.4
|
PFKL
|
phosphofructokinase, liver |
| chr17_-_42908155 | 0.22 |
ENST00000426548.1
ENST00000590758.1 ENST00000591424.1 |
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr22_-_31741757 | 0.22 |
ENST00000215919.3
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr17_+_66287628 | 0.21 |
ENST00000581639.1
ENST00000452479.2 |
ARSG
|
arylsulfatase G |
| chr12_-_15374343 | 0.21 |
ENST00000256953.2
ENST00000546331.1 |
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr12_+_6977258 | 0.21 |
ENST00000488464.2
ENST00000535434.1 ENST00000493987.1 |
TPI1
|
triosephosphate isomerase 1 |
| chr8_-_103424916 | 0.21 |
ENST00000220959.4
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr9_-_115983568 | 0.21 |
ENST00000446284.1
ENST00000414250.1 |
FKBP15
|
FK506 binding protein 15, 133kDa |
| chr14_-_77495007 | 0.20 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr9_-_115983641 | 0.20 |
ENST00000238256.3
|
FKBP15
|
FK506 binding protein 15, 133kDa |
| chr19_+_18530146 | 0.20 |
ENST00000348495.6
ENST00000270061.7 |
SSBP4
|
single stranded DNA binding protein 4 |
| chr16_+_29467127 | 0.20 |
ENST00000344620.6
|
SULT1A4
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr1_-_51425772 | 0.19 |
ENST00000371778.4
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr12_-_64616019 | 0.19 |
ENST00000311915.8
ENST00000398055.3 ENST00000544871.1 |
C12orf66
|
chromosome 12 open reading frame 66 |
| chr2_+_232575128 | 0.19 |
ENST00000412128.1
|
PTMA
|
prothymosin, alpha |
| chr8_+_28351707 | 0.19 |
ENST00000537916.1
ENST00000523546.1 ENST00000240093.3 |
FZD3
|
frizzled family receptor 3 |
| chr3_-_160823158 | 0.19 |
ENST00000392779.2
ENST00000392780.1 ENST00000494173.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr17_-_2206801 | 0.18 |
ENST00000544865.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr19_+_1407517 | 0.18 |
ENST00000336761.6
ENST00000233078.4 |
DAZAP1
|
DAZ associated protein 1 |
| chr8_-_97273807 | 0.18 |
ENST00000517720.1
ENST00000287025.3 ENST00000523821.1 |
MTERFD1
|
MTERF domain containing 1 |
| chr13_+_103451548 | 0.18 |
ENST00000419638.1
|
BIVM
|
basic, immunoglobulin-like variable motif containing |
| chr14_-_39901618 | 0.17 |
ENST00000554932.1
ENST00000298097.7 |
FBXO33
|
F-box protein 33 |
| chr7_-_127032363 | 0.17 |
ENST00000393312.1
|
ZNF800
|
zinc finger protein 800 |
| chr15_-_43622736 | 0.17 |
ENST00000544735.1
ENST00000567039.1 ENST00000305641.5 |
LCMT2
|
leucine carboxyl methyltransferase 2 |
| chr1_-_43833628 | 0.17 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr19_+_50180507 | 0.17 |
ENST00000454376.2
ENST00000524771.1 |
PRMT1
|
protein arginine methyltransferase 1 |
| chr8_+_109455845 | 0.17 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chrX_+_105969893 | 0.17 |
ENST00000255499.2
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr8_+_98656336 | 0.17 |
ENST00000336273.3
|
MTDH
|
metadherin |
| chr11_-_65548265 | 0.16 |
ENST00000532090.2
|
AP5B1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr4_+_4291924 | 0.16 |
ENST00000355834.3
ENST00000337872.4 ENST00000538529.1 ENST00000502918.1 |
ZBTB49
|
zinc finger and BTB domain containing 49 |
| chr12_+_58013693 | 0.16 |
ENST00000320442.4
ENST00000379218.2 |
SLC26A10
|
solute carrier family 26, member 10 |
| chr6_+_3259122 | 0.15 |
ENST00000438998.2
ENST00000380305.4 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr19_+_4472230 | 0.15 |
ENST00000301284.4
ENST00000586684.1 |
HDGFRP2
|
Hepatoma-derived growth factor-related protein 2 |
| chr6_+_151561506 | 0.15 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr15_+_98503922 | 0.15 |
ENST00000268042.6
|
ARRDC4
|
arrestin domain containing 4 |
| chr18_-_45457478 | 0.15 |
ENST00000402690.2
ENST00000356825.4 |
SMAD2
|
SMAD family member 2 |
| chr8_-_103424986 | 0.15 |
ENST00000521922.1
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HIF1A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0034226 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.6 | 2.8 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.4 | 1.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 0.3 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.3 | 0.9 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.3 | 1.6 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.2 | 0.9 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.2 | 0.6 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.2 | 2.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 0.5 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.2 | 0.5 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 1.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.8 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 0.4 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.1 | 0.8 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.5 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.5 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 0.7 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.1 | 0.3 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 1.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.7 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.4 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 0.1 | 0.5 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.1 | 4.5 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.6 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.3 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.1 | 0.5 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 2.1 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 0.6 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.4 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 0.2 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.1 | 0.8 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.6 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.2 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.1 | 1.0 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.3 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) terminal web assembly(GO:1902896) |
| 0.1 | 0.2 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.5 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.3 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0060139 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.3 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.0 | 0.5 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 1.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.5 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.5 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.9 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 1.0 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 1.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) depurination(GO:0045007) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.8 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 2.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.4 | GO:2001225 | regulation of chloride transport(GO:2001225) |
| 0.0 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.3 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.3 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.5 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.0 | 0.3 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 0.1 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.5 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.1 | GO:2000051 | Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.8 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.2 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) |
| 0.0 | 0.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:1904764 | clathrin coat disassembly(GO:0072318) negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.3 | GO:0051481 | stabilization of membrane potential(GO:0030322) negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.0 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.4 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 3.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.2 | 0.8 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 1.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 1.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.4 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.3 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.3 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 1.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 1.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.5 | 2.1 | GO:0005292 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.5 | 3.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 1.2 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.3 | 0.9 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.3 | 0.9 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.3 | 0.8 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.3 | 1.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 0.5 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 0.4 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.4 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.1 | 0.4 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.8 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.6 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 2.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 0.8 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.1 | 0.6 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 0.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.2 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.2 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 1.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.4 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.5 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 1.5 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 3.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |