Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
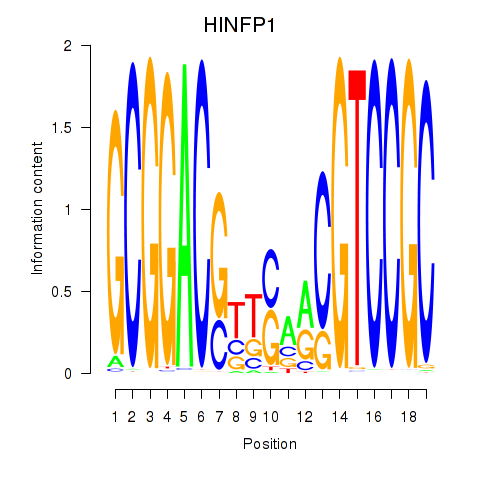
Results for HINFP1
Z-value: 0.51
Transcription factors associated with HINFP1
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of HINFP1 motif
Sorted Z-values of HINFP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_154393260 | 1.53 |
ENST00000435029.4
|
KIF4B
|
kinesin family member 4B |
| chr2_-_234763147 | 1.44 |
ENST00000411486.2
ENST00000432087.1 ENST00000441687.1 ENST00000414924.1 |
HJURP
|
Holliday junction recognition protein |
| chr9_-_94186131 | 1.34 |
ENST00000297689.3
|
NFIL3
|
nuclear factor, interleukin 3 regulated |
| chr18_+_12308046 | 1.28 |
ENST00000317702.5
|
TUBB6
|
tubulin, beta 6 class V |
| chr18_+_12308231 | 1.12 |
ENST00000590103.1
ENST00000591909.1 ENST00000586653.1 ENST00000592683.1 ENST00000590967.1 ENST00000591208.1 ENST00000591463.1 |
TUBB6
|
tubulin, beta 6 class V |
| chr10_-_75634219 | 1.05 |
ENST00000305762.7
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr14_+_55033815 | 1.02 |
ENST00000554335.1
|
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr1_-_94147385 | 1.01 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr10_-_75634260 | 0.98 |
ENST00000372765.1
ENST00000351293.3 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr16_+_57279248 | 0.89 |
ENST00000562023.1
ENST00000563234.1 |
ARL2BP
|
ADP-ribosylation factor-like 2 binding protein |
| chr11_-_61659006 | 0.72 |
ENST00000278829.2
|
FADS3
|
fatty acid desaturase 3 |
| chr10_-_75634326 | 0.71 |
ENST00000322635.3
ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr2_-_130939115 | 0.71 |
ENST00000441135.1
ENST00000339679.7 ENST00000426662.2 ENST00000443958.2 ENST00000351288.6 ENST00000453750.1 ENST00000452225.2 |
SMPD4
|
sphingomyelin phosphodiesterase 4, neutral membrane (neutral sphingomyelinase-3) |
| chr20_+_35974532 | 0.69 |
ENST00000373578.2
|
SRC
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr11_-_61658853 | 0.68 |
ENST00000525588.1
ENST00000540820.1 |
FADS3
|
fatty acid desaturase 3 |
| chr21_+_35445811 | 0.67 |
ENST00000399312.2
|
MRPS6
|
mitochondrial ribosomal protein S6 |
| chr2_+_130939235 | 0.67 |
ENST00000425361.1
ENST00000457492.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr22_+_27053422 | 0.67 |
ENST00000413665.1
ENST00000421151.1 ENST00000456129.1 ENST00000430080.1 |
MIAT
|
myocardial infarction associated transcript (non-protein coding) |
| chr21_+_35445827 | 0.66 |
ENST00000608209.1
ENST00000381151.3 |
SLC5A3
SLC5A3
|
sodium/myo-inositol cotransporter solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
| chr2_+_130939827 | 0.66 |
ENST00000409255.1
ENST00000455239.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr16_+_610407 | 0.65 |
ENST00000409413.3
|
C16orf11
|
chromosome 16 open reading frame 11 |
| chr1_-_43232649 | 0.53 |
ENST00000372526.2
ENST00000236040.4 ENST00000296388.5 ENST00000397054.3 |
LEPRE1
|
leucine proline-enriched proteoglycan (leprecan) 1 |
| chr18_+_11981547 | 0.53 |
ENST00000588927.1
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr3_+_133292851 | 0.52 |
ENST00000503932.1
|
CDV3
|
CDV3 homolog (mouse) |
| chr2_+_58655461 | 0.50 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr3_+_133293278 | 0.48 |
ENST00000508481.1
ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3
|
CDV3 homolog (mouse) |
| chrX_-_125686784 | 0.47 |
ENST00000371126.1
|
DCAF12L1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr1_-_62784935 | 0.44 |
ENST00000354381.3
|
KANK4
|
KN motif and ankyrin repeat domains 4 |
| chr3_+_133292759 | 0.41 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr18_+_11981427 | 0.36 |
ENST00000269159.3
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr13_-_22033392 | 0.28 |
ENST00000320220.9
ENST00000415724.1 ENST00000422251.1 ENST00000382466.3 ENST00000542645.1 ENST00000400590.3 |
ZDHHC20
|
zinc finger, DHHC-type containing 20 |
| chr10_+_72238517 | 0.27 |
ENST00000263563.6
|
PALD1
|
phosphatase domain containing, paladin 1 |
| chrX_-_119249819 | 0.26 |
ENST00000217999.2
|
RHOXF1
|
Rhox homeobox family, member 1 |
| chr15_+_85525205 | 0.24 |
ENST00000394553.1
ENST00000339708.5 |
PDE8A
|
phosphodiesterase 8A |
| chr19_-_14228541 | 0.21 |
ENST00000590853.1
ENST00000308677.4 |
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha |
| chr1_+_222886694 | 0.21 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr1_-_62785054 | 0.19 |
ENST00000371153.4
|
KANK4
|
KN motif and ankyrin repeat domains 4 |
| chr19_-_5680499 | 0.19 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr10_-_977564 | 0.19 |
ENST00000406525.2
|
LARP4B
|
La ribonucleoprotein domain family, member 4B |
| chr1_+_210111534 | 0.18 |
ENST00000422431.1
ENST00000534859.1 ENST00000399639.2 ENST00000537238.1 |
SYT14
|
synaptotagmin XIV |
| chr6_-_144416737 | 0.17 |
ENST00000367569.2
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa |
| chr17_-_79481666 | 0.15 |
ENST00000575659.1
|
ACTG1
|
actin, gamma 1 |
| chr14_+_73704201 | 0.14 |
ENST00000340738.5
ENST00000427855.1 ENST00000381166.3 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr10_-_97453650 | 0.12 |
ENST00000371209.5
ENST00000371217.5 ENST00000430368.2 |
TCTN3
|
tectonic family member 3 |
| chr7_+_6144514 | 0.05 |
ENST00000306177.5
ENST00000465073.2 |
USP42
|
ubiquitin specific peptidase 42 |
| chr1_+_43232913 | 0.04 |
ENST00000372525.5
ENST00000536543.1 |
C1orf50
|
chromosome 1 open reading frame 50 |
| chr1_+_42922173 | 0.02 |
ENST00000455780.1
ENST00000372560.3 ENST00000372561.3 ENST00000372556.3 |
PPCS
|
phosphopantothenoylcysteine synthetase |
Network of associatons between targets according to the STRING database.
First level regulatory network of HINFP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.9 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 2.7 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 0.7 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 1.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 1.4 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 1.0 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.9 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 1.3 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 1.4 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 1.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:1901621 | cellular response to parathyroid hormone stimulus(GO:0071374) negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 2.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.2 | 0.9 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.7 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 1.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.0 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 2.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.7 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 2.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |