Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
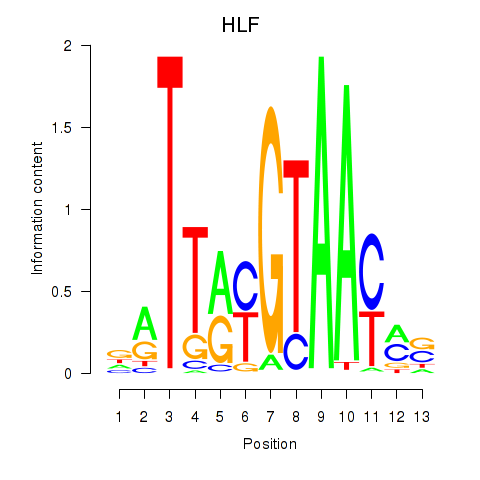
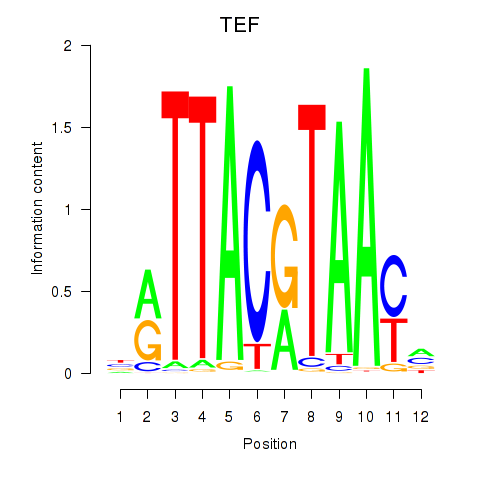
Results for HLF_TEF
Z-value: 0.70
Transcription factors associated with HLF_TEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HLF
|
ENSG00000108924.9 | HLF transcription factor, PAR bZIP family member |
|
TEF
|
ENSG00000167074.10 | TEF transcription factor, PAR bZIP family member |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEF | hg19_v2_chr22_+_41763274_41763337, hg19_v2_chr22_+_41777927_41777971 | 0.34 | 6.4e-02 | Click! |
| HLF | hg19_v2_chr17_+_53344945_53344977 | 0.20 | 2.9e-01 | Click! |
Activity profile of HLF_TEF motif
Sorted Z-values of HLF_TEF motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_33759854 | 1.92 |
ENST00000216968.4
|
PROCR
|
protein C receptor, endothelial |
| chr1_+_7844312 | 1.83 |
ENST00000377541.1
|
PER3
|
period circadian clock 3 |
| chr14_+_78227105 | 1.65 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr2_-_113594279 | 1.61 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr17_-_41623691 | 1.49 |
ENST00000545954.1
|
ETV4
|
ets variant 4 |
| chr17_-_41623075 | 1.41 |
ENST00000545089.1
|
ETV4
|
ets variant 4 |
| chr19_-_35992780 | 1.25 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr17_-_41623259 | 1.12 |
ENST00000538265.1
ENST00000591713.1 |
ETV4
|
ets variant 4 |
| chr2_-_31637560 | 1.10 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr1_-_244013384 | 1.06 |
ENST00000366539.1
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr17_-_41623716 | 1.05 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr5_-_140013275 | 1.05 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr17_-_41623009 | 0.99 |
ENST00000393664.2
|
ETV4
|
ets variant 4 |
| chr12_-_71031185 | 0.98 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr7_-_83824169 | 0.90 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr8_-_6735451 | 0.90 |
ENST00000297439.3
|
DEFB1
|
defensin, beta 1 |
| chr9_+_101705893 | 0.88 |
ENST00000375001.3
|
COL15A1
|
collagen, type XV, alpha 1 |
| chr3_-_48130707 | 0.82 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chr9_-_123639304 | 0.78 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr17_+_73089382 | 0.76 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr3_+_154797877 | 0.71 |
ENST00000462745.1
ENST00000493237.1 |
MME
|
membrane metallo-endopeptidase |
| chr4_+_75230853 | 0.70 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr17_+_39975455 | 0.70 |
ENST00000455106.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr1_+_174933899 | 0.70 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr5_+_150404904 | 0.69 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr2_+_113885138 | 0.68 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr7_+_23145884 | 0.68 |
ENST00000409689.1
ENST00000410047.1 |
KLHL7
|
kelch-like family member 7 |
| chr14_+_96722152 | 0.67 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr1_+_45212051 | 0.65 |
ENST00000372222.3
|
KIF2C
|
kinesin family member 2C |
| chr18_+_32621324 | 0.65 |
ENST00000300249.5
ENST00000538170.2 ENST00000588910.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr1_+_45212074 | 0.63 |
ENST00000372217.1
|
KIF2C
|
kinesin family member 2C |
| chr1_-_226496772 | 0.60 |
ENST00000359525.2
ENST00000460719.1 |
LIN9
|
lin-9 homolog (C. elegans) |
| chr11_+_35201826 | 0.59 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_-_33795893 | 0.57 |
ENST00000526785.1
ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3
|
F-box protein 3 |
| chr16_+_70680439 | 0.56 |
ENST00000288098.2
|
IL34
|
interleukin 34 |
| chr9_-_123639445 | 0.56 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr3_-_48130314 | 0.55 |
ENST00000439356.1
ENST00000395734.3 ENST00000426837.2 |
MAP4
|
microtubule-associated protein 4 |
| chr3_-_99595037 | 0.52 |
ENST00000383694.2
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr9_-_123639600 | 0.52 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr4_-_76928641 | 0.50 |
ENST00000264888.5
|
CXCL9
|
chemokine (C-X-C motif) ligand 9 |
| chr14_+_102276192 | 0.47 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr15_+_81293254 | 0.46 |
ENST00000267984.2
|
MESDC1
|
mesoderm development candidate 1 |
| chr7_+_23146271 | 0.46 |
ENST00000545771.1
|
KLHL7
|
kelch-like family member 7 |
| chr11_-_105948129 | 0.43 |
ENST00000526793.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr9_+_71820057 | 0.42 |
ENST00000539225.1
|
TJP2
|
tight junction protein 2 |
| chr2_+_192543153 | 0.41 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr12_+_93963590 | 0.39 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr1_+_115572415 | 0.39 |
ENST00000256592.1
|
TSHB
|
thyroid stimulating hormone, beta |
| chr6_+_53948328 | 0.39 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr3_-_99594948 | 0.39 |
ENST00000471562.1
ENST00000495625.2 |
FILIP1L
|
filamin A interacting protein 1-like |
| chr4_-_21950356 | 0.39 |
ENST00000447367.2
ENST00000382152.2 |
KCNIP4
|
Kv channel interacting protein 4 |
| chr11_-_62313090 | 0.39 |
ENST00000528508.1
ENST00000533365.1 |
AHNAK
|
AHNAK nucleoprotein |
| chr9_+_71819927 | 0.38 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr4_+_118955500 | 0.38 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr12_-_53074182 | 0.38 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chrX_-_33229636 | 0.38 |
ENST00000357033.4
|
DMD
|
dystrophin |
| chr18_+_21032781 | 0.36 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr11_-_105948040 | 0.36 |
ENST00000534815.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr12_+_57624119 | 0.35 |
ENST00000555773.1
ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr7_+_90032667 | 0.35 |
ENST00000496677.1
ENST00000287916.4 ENST00000535571.1 ENST00000394604.1 ENST00000394605.2 |
CLDN12
|
claudin 12 |
| chr4_+_144257915 | 0.35 |
ENST00000262995.4
|
GAB1
|
GRB2-associated binding protein 1 |
| chr14_+_96722539 | 0.35 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr5_+_95998070 | 0.34 |
ENST00000421689.2
ENST00000510756.1 ENST00000512620.1 |
CAST
|
calpastatin |
| chr10_-_14372870 | 0.34 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr5_+_95997769 | 0.34 |
ENST00000338252.3
ENST00000508830.1 |
CAST
|
calpastatin |
| chr1_-_226496898 | 0.33 |
ENST00000481685.1
|
LIN9
|
lin-9 homolog (C. elegans) |
| chr12_+_57623477 | 0.32 |
ENST00000557487.1
ENST00000555634.1 ENST00000556689.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr12_-_71031220 | 0.32 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr17_-_39538550 | 0.32 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr12_+_57623869 | 0.31 |
ENST00000414700.3
ENST00000557703.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr12_+_57624085 | 0.31 |
ENST00000553474.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr1_-_161207953 | 0.31 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr20_+_37209820 | 0.30 |
ENST00000537425.1
ENST00000373348.3 ENST00000416116.1 |
ADIG
|
adipogenin |
| chr3_+_182983090 | 0.30 |
ENST00000465010.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr9_-_95298314 | 0.30 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr1_-_161208013 | 0.29 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr11_+_69061594 | 0.28 |
ENST00000441339.2
ENST00000308946.3 ENST00000535407.1 |
MYEOV
|
myeloma overexpressed |
| chr3_+_158787041 | 0.27 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr16_-_30102547 | 0.27 |
ENST00000279386.2
|
TBX6
|
T-box 6 |
| chr2_-_101925055 | 0.26 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr13_-_46626847 | 0.26 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr6_-_46459099 | 0.26 |
ENST00000371374.1
|
RCAN2
|
regulator of calcineurin 2 |
| chr1_-_161207986 | 0.25 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr3_-_196910721 | 0.25 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr10_+_94050913 | 0.24 |
ENST00000358935.2
|
MARCH5
|
membrane-associated ring finger (C3HC4) 5 |
| chr16_-_20367584 | 0.24 |
ENST00000570689.1
|
UMOD
|
uromodulin |
| chr8_-_27469196 | 0.24 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr8_+_70404996 | 0.24 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr7_-_37026108 | 0.23 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr1_+_65730385 | 0.23 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr20_+_16729003 | 0.23 |
ENST00000246081.2
|
OTOR
|
otoraplin |
| chr6_-_30043539 | 0.23 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr3_-_58200398 | 0.23 |
ENST00000318316.3
ENST00000460422.1 ENST00000483681.1 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr1_-_114430169 | 0.23 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr14_+_65453432 | 0.23 |
ENST00000246166.2
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr1_-_161207875 | 0.23 |
ENST00000512372.1
ENST00000437437.2 ENST00000442691.2 ENST00000412844.2 ENST00000428574.2 ENST00000505005.1 ENST00000508740.1 ENST00000508387.1 ENST00000504010.1 ENST00000511676.1 ENST00000502985.1 ENST00000367981.3 ENST00000515621.1 ENST00000511944.1 ENST00000511748.1 ENST00000367984.4 ENST00000367985.3 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr5_-_147162263 | 0.22 |
ENST00000333010.6
ENST00000265272.5 |
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr13_-_33780133 | 0.22 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr17_-_39306054 | 0.22 |
ENST00000343246.4
|
KRTAP4-5
|
keratin associated protein 4-5 |
| chr17_-_76124711 | 0.22 |
ENST00000306591.7
ENST00000590602.1 |
TMC6
|
transmembrane channel-like 6 |
| chr19_-_2042065 | 0.22 |
ENST00000591588.1
ENST00000591142.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr12_-_54758251 | 0.21 |
ENST00000267015.3
ENST00000551809.1 |
GPR84
|
G protein-coupled receptor 84 |
| chr6_+_24126350 | 0.21 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr8_+_90914073 | 0.21 |
ENST00000297438.2
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2 |
| chr14_-_72458326 | 0.21 |
ENST00000542853.1
|
AC005477.1
|
AC005477.1 |
| chr17_-_76124812 | 0.21 |
ENST00000592063.1
ENST00000589271.1 ENST00000322933.4 ENST00000589553.1 |
TMC6
|
transmembrane channel-like 6 |
| chr5_-_179050066 | 0.21 |
ENST00000329433.6
ENST00000510411.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr5_-_146435694 | 0.20 |
ENST00000356826.3
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr17_+_11501816 | 0.20 |
ENST00000454412.2
|
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr12_-_75603643 | 0.20 |
ENST00000549446.1
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr1_+_92632542 | 0.20 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr16_+_3507985 | 0.20 |
ENST00000421765.3
ENST00000360862.5 ENST00000414063.2 ENST00000610180.1 ENST00000608993.1 |
NAA60
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit N-alpha-acetyltransferase 60 |
| chr3_+_101504200 | 0.20 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr12_-_88423164 | 0.19 |
ENST00000298699.2
ENST00000550553.1 |
C12orf50
|
chromosome 12 open reading frame 50 |
| chr12_-_75603482 | 0.19 |
ENST00000341669.3
ENST00000298972.1 ENST00000350228.2 |
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr8_+_77318769 | 0.19 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr2_-_201936302 | 0.19 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr16_+_3508063 | 0.18 |
ENST00000576787.1
ENST00000572942.1 ENST00000576916.1 ENST00000575076.1 ENST00000572131.1 |
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr7_-_80141328 | 0.18 |
ENST00000398291.3
|
GNAT3
|
guanine nucleotide binding protein, alpha transducing 3 |
| chr22_+_41487711 | 0.18 |
ENST00000263253.7
|
EP300
|
E1A binding protein p300 |
| chr5_-_114505624 | 0.17 |
ENST00000513154.1
|
TRIM36
|
tripartite motif containing 36 |
| chr5_+_95997918 | 0.17 |
ENST00000395812.2
ENST00000395813.1 ENST00000359176.4 ENST00000325674.7 |
CAST
|
calpastatin |
| chr7_+_20687017 | 0.17 |
ENST00000258738.6
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr1_+_161087873 | 0.17 |
ENST00000368009.2
ENST00000368007.4 ENST00000368008.1 ENST00000392190.5 |
NIT1
|
nitrilase 1 |
| chr4_+_96012614 | 0.17 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr9_+_110045537 | 0.16 |
ENST00000358015.3
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr4_+_144258021 | 0.16 |
ENST00000262994.4
|
GAB1
|
GRB2-associated binding protein 1 |
| chr10_-_82049424 | 0.16 |
ENST00000372213.3
|
MAT1A
|
methionine adenosyltransferase I, alpha |
| chr12_-_120966943 | 0.16 |
ENST00000552443.1
ENST00000547736.1 ENST00000445328.2 ENST00000547943.1 ENST00000288532.6 |
COQ5
|
coenzyme Q5 homolog, methyltransferase (S. cerevisiae) |
| chr1_-_114429997 | 0.15 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr17_-_73851285 | 0.15 |
ENST00000589642.1
ENST00000593002.1 ENST00000590221.1 ENST00000344296.4 ENST00000587374.1 ENST00000585462.1 ENST00000433525.2 ENST00000254806.3 |
WBP2
|
WW domain binding protein 2 |
| chr8_+_68864330 | 0.15 |
ENST00000288368.4
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chrX_+_41306575 | 0.14 |
ENST00000342595.2
ENST00000378220.1 |
NYX
|
nyctalopin |
| chr11_+_62495997 | 0.14 |
ENST00000316461.4
|
TTC9C
|
tetratricopeptide repeat domain 9C |
| chr22_-_29457832 | 0.14 |
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr21_-_30365136 | 0.13 |
ENST00000361371.5
ENST00000389194.2 ENST00000389195.2 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr3_+_148447887 | 0.13 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr1_+_65613513 | 0.13 |
ENST00000395334.2
|
AK4
|
adenylate kinase 4 |
| chr7_+_120629653 | 0.13 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr6_-_31940065 | 0.13 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr5_+_34757309 | 0.12 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr20_-_49547731 | 0.12 |
ENST00000396029.3
|
ADNP
|
activity-dependent neuroprotector homeobox |
| chr8_+_7801144 | 0.12 |
ENST00000443676.1
|
ZNF705B
|
zinc finger protein 705B |
| chr12_-_113574028 | 0.11 |
ENST00000546530.1
ENST00000261729.5 |
RASAL1
|
RAS protein activator like 1 (GAP1 like) |
| chr4_-_96470350 | 0.11 |
ENST00000504962.1
ENST00000453304.1 ENST00000506749.1 |
UNC5C
|
unc-5 homolog C (C. elegans) |
| chr9_-_32552551 | 0.11 |
ENST00000360538.2
ENST00000379858.1 |
TOPORS
|
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase |
| chr22_-_29711704 | 0.11 |
ENST00000216101.6
|
RASL10A
|
RAS-like, family 10, member A |
| chr6_-_64029879 | 0.11 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr14_+_102276132 | 0.11 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr22_-_30867973 | 0.10 |
ENST00000402286.1
ENST00000401751.1 ENST00000539629.1 ENST00000403066.1 ENST00000215812.4 |
SEC14L3
|
SEC14-like 3 (S. cerevisiae) |
| chr20_-_56285595 | 0.10 |
ENST00000395816.3
ENST00000347215.4 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr4_-_153274078 | 0.10 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr1_-_101491319 | 0.09 |
ENST00000342173.7
ENST00000488176.1 ENST00000370109.3 |
DPH5
|
diphthamide biosynthesis 5 |
| chr3_+_157154578 | 0.09 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr19_-_3985455 | 0.09 |
ENST00000309311.6
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr15_+_58430567 | 0.09 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr7_+_135347215 | 0.09 |
ENST00000507606.1
|
C7orf73
|
chromosome 7 open reading frame 73 |
| chr14_+_60715928 | 0.08 |
ENST00000395076.4
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr2_-_112614424 | 0.08 |
ENST00000427997.1
|
ANAPC1
|
anaphase promoting complex subunit 1 |
| chr3_+_186330712 | 0.08 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr15_+_58430368 | 0.08 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr20_-_49547910 | 0.08 |
ENST00000396032.3
|
ADNP
|
activity-dependent neuroprotector homeobox |
| chr13_+_23755099 | 0.08 |
ENST00000537476.1
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr1_+_11249398 | 0.07 |
ENST00000376819.3
|
ANGPTL7
|
angiopoietin-like 7 |
| chr4_+_110354928 | 0.07 |
ENST00000504968.2
ENST00000399100.2 ENST00000265175.5 |
SEC24B
|
SEC24 family member B |
| chr4_+_187187337 | 0.07 |
ENST00000492972.2
|
F11
|
coagulation factor XI |
| chr13_-_21634421 | 0.06 |
ENST00000542899.1
|
LATS2
|
large tumor suppressor kinase 2 |
| chr1_+_75594119 | 0.06 |
ENST00000294638.5
|
LHX8
|
LIM homeobox 8 |
| chr1_-_150552006 | 0.06 |
ENST00000307940.3
ENST00000369026.2 |
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related) |
| chr9_+_27524283 | 0.06 |
ENST00000276943.2
|
IFNK
|
interferon, kappa |
| chr6_+_72926145 | 0.06 |
ENST00000425662.2
ENST00000453976.2 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr7_+_44646162 | 0.06 |
ENST00000439616.2
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr20_+_2082494 | 0.06 |
ENST00000246032.3
|
STK35
|
serine/threonine kinase 35 |
| chr2_+_103378472 | 0.06 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr11_-_72145426 | 0.06 |
ENST00000535990.1
ENST00000437826.2 ENST00000340729.5 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr6_-_116447283 | 0.05 |
ENST00000452729.1
ENST00000243222.4 |
COL10A1
|
collagen, type X, alpha 1 |
| chr8_+_50824233 | 0.05 |
ENST00000522124.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr8_+_42396936 | 0.05 |
ENST00000416469.2
|
SMIM19
|
small integral membrane protein 19 |
| chr8_+_24298597 | 0.05 |
ENST00000380789.1
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr14_+_93260642 | 0.05 |
ENST00000355976.2
|
GOLGA5
|
golgin A5 |
| chr7_+_20686946 | 0.05 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr12_-_68619586 | 0.05 |
ENST00000229134.4
|
IL26
|
interleukin 26 |
| chr10_-_94050820 | 0.05 |
ENST00000265997.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr19_+_50922187 | 0.04 |
ENST00000595883.1
ENST00000597855.1 ENST00000596074.1 ENST00000439922.2 ENST00000594685.1 ENST00000270632.7 |
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr8_+_107738240 | 0.04 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr19_+_33865218 | 0.04 |
ENST00000585933.2
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr10_+_18629628 | 0.04 |
ENST00000377329.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr18_+_77160282 | 0.04 |
ENST00000318065.5
ENST00000545796.1 ENST00000592223.1 ENST00000329101.4 ENST00000586434.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr2_+_191208196 | 0.04 |
ENST00000392329.2
ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr6_+_36238237 | 0.04 |
ENST00000457797.1
ENST00000394571.2 |
PNPLA1
|
patatin-like phospholipase domain containing 1 |
| chr7_+_100210133 | 0.04 |
ENST00000393950.2
ENST00000424091.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr21_+_45138941 | 0.04 |
ENST00000398081.1
ENST00000468090.1 ENST00000291565.4 |
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr2_-_214014959 | 0.04 |
ENST00000442445.1
ENST00000457361.1 ENST00000342002.2 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr8_+_24298531 | 0.04 |
ENST00000175238.6
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr3_-_10547192 | 0.04 |
ENST00000360273.2
ENST00000343816.4 |
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr1_+_154377669 | 0.04 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr1_+_25757376 | 0.03 |
ENST00000399766.3
ENST00000399763.3 ENST00000374343.4 |
TMEM57
|
transmembrane protein 57 |
| chr1_-_72566613 | 0.03 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr19_-_4065730 | 0.03 |
ENST00000601588.1
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr11_-_14993819 | 0.03 |
ENST00000396372.2
ENST00000361010.3 ENST00000359642.3 ENST00000331587.4 |
CALCA
|
calcitonin-related polypeptide alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of HLF_TEF
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.3 | 1.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.3 | 1.0 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.2 | 0.7 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.2 | 0.7 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.2 | 1.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 0.9 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.2 | 1.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.2 | 1.3 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 0.9 | GO:0048880 | sensory system development(GO:0048880) |
| 0.2 | 1.9 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.7 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular response to UV-A(GO:0071492) |
| 0.1 | 1.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.7 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 0.4 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.3 | GO:0070884 | calcineurin-NFAT signaling cascade(GO:0033173) regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 0.8 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.6 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 1.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.5 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.8 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.5 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.8 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.4 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.2 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.3 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.2 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.6 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.2 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 1.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 1.9 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:0061518 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.0 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.5 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.3 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.2 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.0 | 0.2 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.4 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.2 | GO:0001502 | cartilage condensation(GO:0001502) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:1990742 | microvesicle(GO:1990742) |
| 0.2 | 1.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 1.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.9 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.6 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 1.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 1.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.1 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.3 | 0.9 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 0.7 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.2 | 1.0 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 1.3 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.2 | 1.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 1.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.3 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 1.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 1.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 1.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.0 | GO:0031403 | pyridoxal kinase activity(GO:0008478) lithium ion binding(GO:0031403) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 6.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.0 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 6.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 2.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |