Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
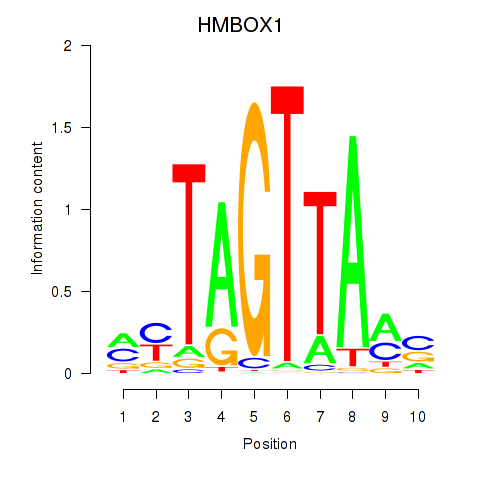
Results for HMBOX1
Z-value: 0.82
Transcription factors associated with HMBOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMBOX1
|
ENSG00000147421.13 | homeobox containing 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMBOX1 | hg19_v2_chr8_+_28747884_28747938 | 0.63 | 1.8e-04 | Click! |
Activity profile of HMBOX1 motif
Sorted Z-values of HMBOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_25348007 | 4.35 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr6_+_88117683 | 3.77 |
ENST00000369562.4
|
C6ORF165
|
UPF0704 protein C6orf165 |
| chr6_+_112408768 | 3.45 |
ENST00000368656.2
ENST00000604268.1 |
FAM229B
|
family with sequence similarity 229, member B |
| chr9_-_117111222 | 2.58 |
ENST00000374079.4
|
AKNA
|
AT-hook transcription factor |
| chr4_+_9172135 | 2.29 |
ENST00000512047.1
|
FAM90A26
|
family with sequence similarity 90, member A26 |
| chr6_-_52668605 | 2.21 |
ENST00000334575.5
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr13_+_24144509 | 2.10 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr14_+_75536335 | 1.97 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr14_+_75536280 | 1.92 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr1_-_169396666 | 1.83 |
ENST00000456107.1
ENST00000367805.3 |
CCDC181
|
coiled-coil domain containing 181 |
| chr21_-_46340884 | 1.80 |
ENST00000302347.5
ENST00000517819.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr2_+_61293021 | 1.80 |
ENST00000402291.1
|
KIAA1841
|
KIAA1841 |
| chr20_+_43211149 | 1.69 |
ENST00000372886.1
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr1_-_169396646 | 1.68 |
ENST00000367806.3
|
CCDC181
|
coiled-coil domain containing 181 |
| chr1_+_174844645 | 1.64 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr8_+_67976593 | 1.62 |
ENST00000262210.5
ENST00000412460.1 |
CSPP1
|
centrosome and spindle pole associated protein 1 |
| chr3_-_57530051 | 1.61 |
ENST00000311202.6
ENST00000351747.2 ENST00000495027.1 ENST00000389536.4 |
DNAH12
|
dynein, axonemal, heavy chain 12 |
| chr2_-_220110187 | 1.53 |
ENST00000295759.7
ENST00000392089.2 |
GLB1L
|
galactosidase, beta 1-like |
| chr2_-_220110111 | 1.53 |
ENST00000428427.1
ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L
|
galactosidase, beta 1-like |
| chrX_-_10851762 | 1.52 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr2_-_62081148 | 1.51 |
ENST00000404929.1
|
FAM161A
|
family with sequence similarity 161, member A |
| chr2_-_62081254 | 1.50 |
ENST00000405894.3
|
FAM161A
|
family with sequence similarity 161, member A |
| chr4_+_36283213 | 1.41 |
ENST00000357504.3
|
DTHD1
|
death domain containing 1 |
| chr12_-_56236734 | 1.40 |
ENST00000548629.1
|
MMP19
|
matrix metallopeptidase 19 |
| chr12_-_113658892 | 1.38 |
ENST00000299732.2
ENST00000416617.2 |
IQCD
|
IQ motif containing D |
| chr7_-_120498357 | 1.36 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr22_-_37172111 | 1.27 |
ENST00000417951.2
ENST00000430701.1 ENST00000433985.2 |
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr11_+_100862811 | 1.25 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr7_+_100728720 | 1.23 |
ENST00000306085.6
ENST00000412507.1 |
TRIM56
|
tripartite motif containing 56 |
| chr22_-_37172191 | 1.23 |
ENST00000340630.5
|
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr18_-_53253323 | 1.20 |
ENST00000540999.1
ENST00000563888.2 |
TCF4
|
transcription factor 4 |
| chr14_+_45431379 | 1.15 |
ENST00000361577.3
ENST00000361462.2 ENST00000382233.2 |
FAM179B
|
family with sequence similarity 179, member B |
| chr17_+_26800648 | 1.13 |
ENST00000545060.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr1_-_217311090 | 1.13 |
ENST00000493603.1
ENST00000366940.2 |
ESRRG
|
estrogen-related receptor gamma |
| chr17_+_26800756 | 1.12 |
ENST00000537681.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr8_-_28347737 | 1.10 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr6_+_30951487 | 1.09 |
ENST00000486149.2
ENST00000376296.3 |
MUC21
|
mucin 21, cell surface associated |
| chr1_-_86861660 | 1.07 |
ENST00000486215.1
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chrX_+_16964985 | 1.03 |
ENST00000303843.7
|
REPS2
|
RALBP1 associated Eps domain containing 2 |
| chr6_-_52710893 | 1.02 |
ENST00000284562.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr17_+_26800296 | 0.99 |
ENST00000444914.3
ENST00000314669.5 |
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr10_-_112678904 | 0.96 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr1_-_150669500 | 0.93 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr4_-_110723134 | 0.91 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr1_+_153940713 | 0.90 |
ENST00000368601.1
ENST00000368603.1 ENST00000368600.3 |
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr1_-_217250231 | 0.89 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr4_-_110723194 | 0.89 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr14_+_50999744 | 0.85 |
ENST00000441560.2
|
ATL1
|
atlastin GTPase 1 |
| chr14_-_89878369 | 0.85 |
ENST00000553840.1
ENST00000556916.1 |
FOXN3
|
forkhead box N3 |
| chrX_-_10588459 | 0.81 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr5_+_140254884 | 0.77 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chr1_+_27561007 | 0.77 |
ENST00000319394.3
|
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr6_+_27114861 | 0.75 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr6_-_165989936 | 0.74 |
ENST00000354448.4
|
PDE10A
|
phosphodiesterase 10A |
| chr6_-_52628271 | 0.73 |
ENST00000493422.1
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr1_-_86861936 | 0.69 |
ENST00000394733.2
ENST00000359242.3 ENST00000294678.2 ENST00000479890.1 ENST00000317336.7 ENST00000370567.1 ENST00000394731.1 ENST00000478286.2 ENST00000370566.3 |
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chrX_-_10588595 | 0.68 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr6_+_46097711 | 0.66 |
ENST00000321037.4
|
ENPP4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative) |
| chr17_+_48172639 | 0.65 |
ENST00000503176.1
ENST00000503614.1 |
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2 |
| chrX_-_115594160 | 0.63 |
ENST00000371894.4
|
CXorf61
|
cancer/testis antigen 83 |
| chr6_-_15586238 | 0.63 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr2_+_24714729 | 0.62 |
ENST00000406961.1
ENST00000405141.1 |
NCOA1
|
nuclear receptor coactivator 1 |
| chr15_+_75639296 | 0.57 |
ENST00000564500.1
ENST00000355059.4 ENST00000566752.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr3_-_69101461 | 0.56 |
ENST00000543976.1
|
TMF1
|
TATA element modulatory factor 1 |
| chr1_+_109255279 | 0.56 |
ENST00000370017.3
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr3_-_69101413 | 0.56 |
ENST00000398559.2
|
TMF1
|
TATA element modulatory factor 1 |
| chr16_-_30597000 | 0.55 |
ENST00000470110.1
ENST00000395216.2 |
ZNF785
|
zinc finger protein 785 |
| chr2_-_197226875 | 0.54 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr7_-_47579188 | 0.53 |
ENST00000398879.1
ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3
|
tensin 3 |
| chr3_+_52444651 | 0.52 |
ENST00000327906.3
|
PHF7
|
PHD finger protein 7 |
| chr1_+_222988464 | 0.52 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr1_+_222988363 | 0.50 |
ENST00000450784.1
ENST00000426045.1 ENST00000457955.1 ENST00000444858.1 ENST00000435378.1 ENST00000441676.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr12_-_56727676 | 0.48 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr4_+_25915896 | 0.46 |
ENST00000514384.1
|
SMIM20
|
small integral membrane protein 20 |
| chr4_+_25915822 | 0.46 |
ENST00000506197.2
|
SMIM20
|
small integral membrane protein 20 |
| chr12_+_94071341 | 0.45 |
ENST00000542893.2
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr10_-_112678976 | 0.45 |
ENST00000448814.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr11_+_19798964 | 0.44 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr1_+_27561104 | 0.44 |
ENST00000361771.3
|
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr2_+_128458514 | 0.43 |
ENST00000310981.4
|
SFT2D3
|
SFT2 domain containing 3 |
| chr8_+_27491572 | 0.43 |
ENST00000301904.3
|
SCARA3
|
scavenger receptor class A, member 3 |
| chr3_+_69928256 | 0.43 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr2_+_127413481 | 0.43 |
ENST00000259254.4
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr17_-_56609302 | 0.43 |
ENST00000581607.1
ENST00000317256.6 ENST00000426861.1 ENST00000580809.1 ENST00000577729.1 ENST00000583291.1 |
SEPT4
|
septin 4 |
| chr17_-_29641084 | 0.43 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chrX_+_70586140 | 0.42 |
ENST00000276072.3
|
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chrX_+_70752917 | 0.41 |
ENST00000373719.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr5_+_131705438 | 0.41 |
ENST00000245407.3
|
SLC22A5
|
solute carrier family 22 (organic cation/carnitine transporter), member 5 |
| chr20_+_42187682 | 0.40 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr12_-_56727487 | 0.39 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr11_-_5207612 | 0.39 |
ENST00000380367.1
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1 |
| chr10_+_24755416 | 0.38 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr17_-_62493131 | 0.38 |
ENST00000539111.2
|
POLG2
|
polymerase (DNA directed), gamma 2, accessory subunit |
| chr1_-_93257951 | 0.37 |
ENST00000543509.1
ENST00000370331.1 ENST00000540033.1 |
EVI5
|
ecotropic viral integration site 5 |
| chr8_+_120885949 | 0.37 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr1_+_109632425 | 0.36 |
ENST00000338272.8
|
TMEM167B
|
transmembrane protein 167B |
| chr9_+_33290491 | 0.36 |
ENST00000379540.3
ENST00000379521.4 ENST00000318524.6 |
NFX1
|
nuclear transcription factor, X-box binding 1 |
| chr1_+_206643787 | 0.36 |
ENST00000367120.3
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr19_-_7553852 | 0.36 |
ENST00000593547.1
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma |
| chr14_+_56078695 | 0.35 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr14_-_23755297 | 0.35 |
ENST00000357460.5
|
HOMEZ
|
homeobox and leucine zipper encoding |
| chr12_+_104982622 | 0.33 |
ENST00000549016.1
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr6_-_112408661 | 0.33 |
ENST00000368662.5
|
TUBE1
|
tubulin, epsilon 1 |
| chr2_+_68384976 | 0.33 |
ENST00000263657.2
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae) |
| chr12_-_54673871 | 0.33 |
ENST00000209875.4
|
CBX5
|
chromobox homolog 5 |
| chr5_+_178977546 | 0.33 |
ENST00000319449.4
ENST00000377001.2 |
RUFY1
|
RUN and FYVE domain containing 1 |
| chrX_+_22291065 | 0.32 |
ENST00000323684.1
|
ZNF645
|
zinc finger protein 645 |
| chr17_-_27621125 | 0.31 |
ENST00000579665.1
ENST00000225388.4 |
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr10_+_89419370 | 0.31 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chrX_-_99987088 | 0.31 |
ENST00000372981.1
ENST00000263033.5 |
SYTL4
|
synaptotagmin-like 4 |
| chr13_+_111365602 | 0.31 |
ENST00000333219.7
|
ING1
|
inhibitor of growth family, member 1 |
| chr18_-_19283649 | 0.30 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr5_+_173316341 | 0.30 |
ENST00000520867.1
ENST00000334035.5 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr3_+_38347427 | 0.30 |
ENST00000273173.4
|
SLC22A14
|
solute carrier family 22, member 14 |
| chr14_+_105331596 | 0.30 |
ENST00000556508.1
ENST00000414716.3 ENST00000453495.1 ENST00000418279.1 |
CEP170B
|
centrosomal protein 170B |
| chr1_+_151043070 | 0.30 |
ENST00000368918.3
ENST00000368917.1 |
GABPB2
|
GA binding protein transcription factor, beta subunit 2 |
| chr20_+_42187608 | 0.30 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr19_-_7553889 | 0.30 |
ENST00000221480.1
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma |
| chr5_+_110559603 | 0.29 |
ENST00000512453.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr2_+_109335929 | 0.29 |
ENST00000283195.6
|
RANBP2
|
RAN binding protein 2 |
| chr10_+_115312766 | 0.29 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr11_-_124981475 | 0.28 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr4_-_186877502 | 0.28 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr19_+_41699135 | 0.28 |
ENST00000542619.1
ENST00000600561.1 |
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr1_-_159046617 | 0.27 |
ENST00000368130.4
|
AIM2
|
absent in melanoma 2 |
| chr18_-_32870148 | 0.27 |
ENST00000589178.1
ENST00000333206.5 ENST00000592278.1 ENST00000592211.1 ENST00000420878.3 ENST00000383091.2 ENST00000586922.2 |
ZSCAN30
RP11-158H5.7
|
zinc finger and SCAN domain containing 30 RP11-158H5.7 |
| chr1_+_198126093 | 0.26 |
ENST00000367385.4
ENST00000442588.1 ENST00000538004.1 |
NEK7
|
NIMA-related kinase 7 |
| chr6_-_26199471 | 0.26 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr9_-_101984184 | 0.26 |
ENST00000476832.1
|
ALG2
|
ALG2, alpha-1,3/1,6-mannosyltransferase |
| chr3_+_37035289 | 0.25 |
ENST00000455445.2
ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1
|
mutL homolog 1 |
| chr1_+_196743943 | 0.25 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr1_+_100436065 | 0.24 |
ENST00000370153.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr14_+_32030582 | 0.24 |
ENST00000550649.1
ENST00000281081.7 |
NUBPL
|
nucleotide binding protein-like |
| chr8_-_38326139 | 0.24 |
ENST00000335922.5
ENST00000532791.1 ENST00000397091.5 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr19_-_53238260 | 0.24 |
ENST00000453741.2
ENST00000602162.1 ENST00000601643.1 ENST00000596702.1 ENST00000600943.1 ENST00000543227.1 ENST00000540744.1 |
ZNF611
|
zinc finger protein 611 |
| chr20_-_54580523 | 0.24 |
ENST00000064571.2
|
CBLN4
|
cerebellin 4 precursor |
| chr11_-_59950519 | 0.23 |
ENST00000528851.1
|
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr8_-_95220775 | 0.23 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr11_-_47447970 | 0.23 |
ENST00000298852.3
ENST00000530912.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr11_+_17281900 | 0.23 |
ENST00000530527.1
|
NUCB2
|
nucleobindin 2 |
| chrX_+_70586082 | 0.23 |
ENST00000373790.4
ENST00000449580.1 ENST00000423759.1 |
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr11_-_62572901 | 0.23 |
ENST00000439713.2
ENST00000531131.1 ENST00000530875.1 ENST00000531709.2 ENST00000294172.2 |
NXF1
|
nuclear RNA export factor 1 |
| chr3_-_151176497 | 0.23 |
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr11_-_59950486 | 0.22 |
ENST00000426738.2
ENST00000533023.1 ENST00000420732.2 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr7_-_22234381 | 0.22 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr12_+_51633061 | 0.22 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr14_+_31028329 | 0.21 |
ENST00000206595.6
|
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr11_-_46638720 | 0.20 |
ENST00000326737.3
|
HARBI1
|
harbinger transposase derived 1 |
| chrM_+_5824 | 0.20 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr1_-_160549235 | 0.19 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr3_-_12587055 | 0.19 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr12_-_122018859 | 0.18 |
ENST00000536437.1
ENST00000377071.4 ENST00000538046.2 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr12_+_49121213 | 0.17 |
ENST00000548380.1
|
LINC00935
|
long intergenic non-protein coding RNA 935 |
| chr1_+_196743912 | 0.17 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr12_+_4758264 | 0.17 |
ENST00000266544.5
|
NDUFA9
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
| chrX_+_70752945 | 0.17 |
ENST00000373701.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr3_-_45837959 | 0.16 |
ENST00000353278.4
ENST00000456124.2 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr12_-_53715328 | 0.16 |
ENST00000547757.1
ENST00000394384.3 ENST00000209873.4 |
AAAS
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr10_+_35484793 | 0.16 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr6_+_30585486 | 0.16 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr14_+_31028348 | 0.16 |
ENST00000550944.1
ENST00000438909.2 ENST00000553504.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr8_-_116504448 | 0.15 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr5_-_133968459 | 0.15 |
ENST00000505758.1
ENST00000439578.1 ENST00000502286.1 |
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr11_-_111649015 | 0.15 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr17_-_55822653 | 0.14 |
ENST00000299415.2
|
AC007431.1
|
coiled-coil domain containing 182 |
| chr17_-_29641104 | 0.14 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr6_+_29364416 | 0.14 |
ENST00000383555.2
|
OR12D2
|
olfactory receptor, family 12, subfamily D, member 2 (gene/pseudogene) |
| chr19_+_57019212 | 0.14 |
ENST00000308031.5
ENST00000591537.1 |
ZNF471
|
zinc finger protein 471 |
| chr11_+_34643600 | 0.14 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr2_+_106468204 | 0.13 |
ENST00000425756.1
ENST00000393349.2 |
NCK2
|
NCK adaptor protein 2 |
| chr5_-_39074479 | 0.13 |
ENST00000514735.1
ENST00000296782.5 ENST00000357387.3 |
RICTOR
|
RPTOR independent companion of MTOR, complex 2 |
| chr14_+_37641012 | 0.13 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chr17_+_3118915 | 0.13 |
ENST00000304094.1
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1 |
| chr8_+_107738240 | 0.13 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr6_-_26199499 | 0.13 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr11_+_10772534 | 0.13 |
ENST00000361367.2
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr4_-_69434245 | 0.13 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr1_+_206643806 | 0.13 |
ENST00000537984.1
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr4_-_47465666 | 0.13 |
ENST00000381571.4
|
COMMD8
|
COMM domain containing 8 |
| chr9_-_114246635 | 0.13 |
ENST00000338205.5
|
KIAA0368
|
KIAA0368 |
| chr7_+_101460882 | 0.13 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr19_-_56826157 | 0.12 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr3_-_57326704 | 0.12 |
ENST00000487349.1
ENST00000389601.3 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr14_-_45431091 | 0.12 |
ENST00000579157.1
ENST00000396128.4 ENST00000556500.1 |
KLHL28
|
kelch-like family member 28 |
| chr2_-_3521518 | 0.12 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chrX_+_135730373 | 0.11 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chrX_+_66764375 | 0.11 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr1_-_149982624 | 0.11 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chrX_+_135730297 | 0.11 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr1_+_175036966 | 0.11 |
ENST00000239462.4
|
TNN
|
tenascin N |
| chr5_+_138629417 | 0.11 |
ENST00000510056.1
ENST00000511249.1 ENST00000503811.1 ENST00000511378.1 |
MATR3
|
matrin 3 |
| chr2_-_86948245 | 0.11 |
ENST00000439940.2
ENST00000604011.1 |
CHMP3
RNF103-CHMP3
|
charged multivesicular body protein 3 RNF103-CHMP3 readthrough |
| chr19_+_50084561 | 0.11 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr3_-_126327398 | 0.11 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr1_+_24286287 | 0.10 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr12_+_28343365 | 0.10 |
ENST00000545336.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr19_+_50353944 | 0.09 |
ENST00000594151.1
ENST00000600603.1 ENST00000601638.1 ENST00000221557.9 |
PTOV1
|
prostate tumor overexpressed 1 |
| chr13_-_77901177 | 0.09 |
ENST00000407578.2
ENST00000357337.6 ENST00000360084.5 |
MYCBP2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr13_-_67802549 | 0.09 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr1_-_154832316 | 0.09 |
ENST00000361147.4
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMBOX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:2000845 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.2 | 0.9 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.2 | 3.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 0.6 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.2 | 1.4 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 1.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.6 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.1 | 2.2 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.1 | 0.7 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.4 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 1.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.2 | GO:0051257 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 1.0 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.2 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 2.5 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.1 | 0.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 1.5 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.2 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 0.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 1.2 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.6 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.4 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.2 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.4 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.1 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.4 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.6 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.9 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 1.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.8 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 2.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.7 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 1.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.7 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 1.2 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.4 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 2.7 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 1.0 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.3 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.5 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.0 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.7 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.8 | GO:0001942 | hair follicle development(GO:0001942) |
| 0.0 | 1.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.3 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 1.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.5 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 1.8 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.2 | 0.9 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 0.6 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.4 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.1 | 1.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 2.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.2 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.1 | 0.8 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 3.0 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.4 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.6 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 2.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 2.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.3 | 3.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.3 | 2.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 1.8 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.2 | 0.5 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.2 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.6 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.6 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.1 | 0.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.3 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 0.3 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 1.2 | GO:0001083 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.4 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.2 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 2.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 1.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.3 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 1.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.6 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 1.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 1.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 2.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 2.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 4.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |