Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for HMX1
Z-value: 0.64
Transcription factors associated with HMX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX1
|
ENSG00000215612.5 | H6 family homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMX1 | hg19_v2_chr4_-_8873531_8873543 | 0.21 | 2.6e-01 | Click! |
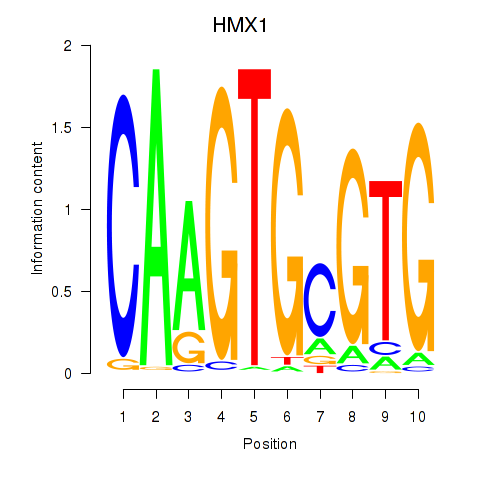
Activity profile of HMX1 motif
Sorted Z-values of HMX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_21494654 | 2.16 |
ENST00000377142.4
|
NKX2-2
|
NK2 homeobox 2 |
| chr2_+_106682103 | 2.09 |
ENST00000238044.3
|
C2orf40
|
chromosome 2 open reading frame 40 |
| chr2_+_106682135 | 1.73 |
ENST00000437659.1
|
C2orf40
|
chromosome 2 open reading frame 40 |
| chr16_-_776431 | 1.42 |
ENST00000293889.6
|
CCDC78
|
coiled-coil domain containing 78 |
| chr18_-_45935663 | 1.16 |
ENST00000589194.1
ENST00000591279.1 ENST00000590855.1 ENST00000587107.1 ENST00000588970.1 ENST00000586525.1 ENST00000592387.1 ENST00000590800.1 |
ZBTB7C
|
zinc finger and BTB domain containing 7C |
| chr1_-_46598371 | 1.10 |
ENST00000372006.1
ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr1_-_46598284 | 1.08 |
ENST00000423209.1
ENST00000262741.5 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr20_+_44098346 | 1.04 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr13_+_25254545 | 1.03 |
ENST00000218548.6
|
ATP12A
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide |
| chr5_+_156693159 | 0.98 |
ENST00000347377.6
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr5_+_156693091 | 0.95 |
ENST00000318218.6
ENST00000442283.2 ENST00000522463.1 ENST00000521420.1 |
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr13_+_35516390 | 0.94 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr19_-_14316980 | 0.92 |
ENST00000361434.3
ENST00000340736.6 |
LPHN1
|
latrophilin 1 |
| chr3_-_107941209 | 0.84 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr3_+_69788576 | 0.80 |
ENST00000352241.4
ENST00000448226.2 |
MITF
|
microphthalmia-associated transcription factor |
| chr22_-_18923655 | 0.74 |
ENST00000438924.1
ENST00000457083.1 ENST00000420436.1 ENST00000334029.2 ENST00000357068.6 |
PRODH
|
proline dehydrogenase (oxidase) 1 |
| chr13_+_25254693 | 0.73 |
ENST00000381946.3
|
ATP12A
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide |
| chr2_-_27712583 | 0.72 |
ENST00000260570.3
ENST00000359466.6 ENST00000416524.2 |
IFT172
|
intraflagellar transport 172 homolog (Chlamydomonas) |
| chr11_-_110561721 | 0.68 |
ENST00000357139.3
|
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr22_-_29196030 | 0.68 |
ENST00000405219.3
|
XBP1
|
X-box binding protein 1 |
| chr17_-_72709050 | 0.64 |
ENST00000583937.1
ENST00000301573.9 ENST00000326165.6 ENST00000469092.1 |
CD300LF
|
CD300 molecule-like family member f |
| chr16_-_70712229 | 0.61 |
ENST00000562883.2
|
MTSS1L
|
metastasis suppressor 1-like |
| chr1_-_177133818 | 0.61 |
ENST00000424564.2
ENST00000361833.2 |
ASTN1
|
astrotactin 1 |
| chr5_-_81046922 | 0.60 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr2_-_46769694 | 0.60 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr22_-_29196546 | 0.58 |
ENST00000403532.3
ENST00000216037.6 |
XBP1
|
X-box binding protein 1 |
| chr20_+_44098385 | 0.56 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr17_-_6947225 | 0.54 |
ENST00000574600.1
ENST00000308009.1 ENST00000447225.1 |
SLC16A11
|
solute carrier family 16, member 11 |
| chr19_+_41305330 | 0.54 |
ENST00000593972.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr9_+_976964 | 0.54 |
ENST00000190165.2
|
DMRT3
|
doublesex and mab-3 related transcription factor 3 |
| chr1_-_110052302 | 0.52 |
ENST00000369864.4
ENST00000369862.1 |
AMIGO1
|
adhesion molecule with Ig-like domain 1 |
| chr12_-_15374343 | 0.52 |
ENST00000256953.2
ENST00000546331.1 |
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr19_+_41305627 | 0.51 |
ENST00000593525.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr3_-_107941230 | 0.51 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr8_-_17270809 | 0.51 |
ENST00000180173.5
ENST00000521857.1 |
MTMR7
|
myotubularin related protein 7 |
| chr19_+_41305740 | 0.50 |
ENST00000596517.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr19_+_41305612 | 0.49 |
ENST00000594380.1
ENST00000593397.1 ENST00000601733.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr2_-_233352531 | 0.49 |
ENST00000304546.1
|
ECEL1
|
endothelin converting enzyme-like 1 |
| chr14_+_56585048 | 0.49 |
ENST00000267460.4
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr17_-_72968809 | 0.49 |
ENST00000530857.1
ENST00000425042.2 |
HID1
|
HID1 domain containing |
| chr16_+_30210552 | 0.48 |
ENST00000338971.5
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr12_+_8234807 | 0.48 |
ENST00000339754.5
|
NECAP1
|
NECAP endocytosis associated 1 |
| chr16_+_30212378 | 0.47 |
ENST00000569485.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr16_+_28996364 | 0.47 |
ENST00000564277.1
|
LAT
|
linker for activation of T cells |
| chr12_+_41831485 | 0.46 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr1_+_9648921 | 0.46 |
ENST00000377376.4
ENST00000340305.5 ENST00000340381.6 |
TMEM201
|
transmembrane protein 201 |
| chr19_+_41305085 | 0.46 |
ENST00000303961.4
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr11_+_6502675 | 0.45 |
ENST00000254616.6
ENST00000530751.1 |
TIMM10B
|
translocase of inner mitochondrial membrane 10 homolog B (yeast) |
| chr4_-_141348789 | 0.44 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr7_-_19184929 | 0.44 |
ENST00000275461.3
|
FERD3L
|
Fer3-like bHLH transcription factor |
| chr19_+_41305406 | 0.43 |
ENST00000406058.2
ENST00000593726.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr4_-_141348999 | 0.42 |
ENST00000325617.5
|
CLGN
|
calmegin |
| chr19_+_38085731 | 0.42 |
ENST00000589117.1
|
ZNF540
|
zinc finger protein 540 |
| chr7_-_37488834 | 0.41 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr17_-_47755436 | 0.41 |
ENST00000505581.1
ENST00000514121.1 ENST00000393328.2 ENST00000509079.1 ENST00000393331.3 ENST00000347630.2 ENST00000504102.1 |
SPOP
|
speckle-type POZ protein |
| chr1_+_156338993 | 0.39 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr13_-_97646604 | 0.39 |
ENST00000298440.1
ENST00000543457.1 ENST00000541038.1 |
OXGR1
|
oxoglutarate (alpha-ketoglutarate) receptor 1 |
| chr20_-_3140490 | 0.38 |
ENST00000449731.1
ENST00000380266.3 |
UBOX5
FASTKD5
|
U-box domain containing 5 FAST kinase domains 5 |
| chr7_-_105332084 | 0.38 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr16_+_29472707 | 0.37 |
ENST00000565290.1
|
SULT1A4
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr1_-_217311090 | 0.37 |
ENST00000493603.1
ENST00000366940.2 |
ESRRG
|
estrogen-related receptor gamma |
| chr5_-_81046841 | 0.36 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr16_-_70713928 | 0.36 |
ENST00000576338.1
|
MTSS1L
|
metastasis suppressor 1-like |
| chr3_-_192445289 | 0.36 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr8_+_145726472 | 0.36 |
ENST00000528430.1
|
PPP1R16A
|
protein phosphatase 1, regulatory subunit 16A |
| chr15_-_58306295 | 0.36 |
ENST00000559517.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr15_+_33010175 | 0.34 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr5_-_81046904 | 0.34 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr17_-_27278445 | 0.34 |
ENST00000268756.3
ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr1_-_207095324 | 0.33 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr19_-_45579762 | 0.33 |
ENST00000303809.2
|
ZNF296
|
zinc finger protein 296 |
| chr20_-_22559211 | 0.33 |
ENST00000564492.1
|
LINC00261
|
long intergenic non-protein coding RNA 261 |
| chr2_+_204801471 | 0.33 |
ENST00000316386.6
ENST00000435193.1 |
ICOS
|
inducible T-cell co-stimulator |
| chr17_-_27278304 | 0.32 |
ENST00000577226.1
|
PHF12
|
PHD finger protein 12 |
| chr11_+_66624527 | 0.32 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr18_-_47721447 | 0.32 |
ENST00000285039.7
|
MYO5B
|
myosin VB |
| chr17_+_40440481 | 0.32 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr5_+_10353780 | 0.32 |
ENST00000449913.2
ENST00000503788.1 ENST00000274140.5 |
MARCH6
|
membrane-associated ring finger (C3HC4) 6, E3 ubiquitin protein ligase |
| chr16_+_67226019 | 0.31 |
ENST00000379378.3
|
E2F4
|
E2F transcription factor 4, p107/p130-binding |
| chr4_-_122854193 | 0.31 |
ENST00000513531.1
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr9_+_100000717 | 0.31 |
ENST00000375205.2
ENST00000357054.1 ENST00000395220.1 ENST00000375202.2 ENST00000411667.2 |
CCDC180
|
coiled-coil domain containing 180 |
| chr11_+_1411503 | 0.29 |
ENST00000526678.1
|
BRSK2
|
BR serine/threonine kinase 2 |
| chr16_+_56385290 | 0.29 |
ENST00000564727.1
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr7_-_156685841 | 0.29 |
ENST00000354505.4
ENST00000540390.1 |
LMBR1
|
limb development membrane protein 1 |
| chrX_+_13587712 | 0.29 |
ENST00000361306.1
ENST00000380602.3 |
EGFL6
|
EGF-like-domain, multiple 6 |
| chr8_-_75233563 | 0.27 |
ENST00000342232.4
|
JPH1
|
junctophilin 1 |
| chr22_-_39268192 | 0.27 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr22_-_38349552 | 0.27 |
ENST00000422191.1
ENST00000249079.2 ENST00000418863.1 ENST00000403305.1 ENST00000403026.1 |
C22orf23
|
chromosome 22 open reading frame 23 |
| chr17_-_62097904 | 0.27 |
ENST00000583366.1
|
ICAM2
|
intercellular adhesion molecule 2 |
| chr18_-_56940611 | 0.27 |
ENST00000256852.7
ENST00000334889.3 |
RAX
|
retina and anterior neural fold homeobox |
| chr9_-_140082983 | 0.26 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr6_-_2245892 | 0.25 |
ENST00000380815.4
|
GMDS
|
GDP-mannose 4,6-dehydratase |
| chr20_-_35374456 | 0.25 |
ENST00000373803.2
ENST00000359675.2 ENST00000540765.1 ENST00000349004.1 |
NDRG3
|
NDRG family member 3 |
| chr21_-_40685504 | 0.25 |
ENST00000380800.3
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr1_-_28241226 | 0.24 |
ENST00000373912.3
ENST00000373909.3 |
RPA2
|
replication protein A2, 32kDa |
| chr1_-_207095212 | 0.23 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr12_+_125478241 | 0.23 |
ENST00000341446.8
|
BRI3BP
|
BRI3 binding protein |
| chr7_-_127032363 | 0.23 |
ENST00000393312.1
|
ZNF800
|
zinc finger protein 800 |
| chr16_+_28996416 | 0.23 |
ENST00000395456.2
ENST00000454369.2 |
LAT
|
linker for activation of T cells |
| chr1_-_28241024 | 0.23 |
ENST00000313433.7
ENST00000444045.1 |
RPA2
|
replication protein A2, 32kDa |
| chr17_-_42298201 | 0.23 |
ENST00000527034.1
|
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr12_+_70133152 | 0.22 |
ENST00000550536.1
ENST00000362025.5 |
RAB3IP
|
RAB3A interacting protein |
| chr16_+_19535133 | 0.22 |
ENST00000396212.2
ENST00000381396.5 |
CCP110
|
centriolar coiled coil protein 110kDa |
| chr16_+_19535235 | 0.22 |
ENST00000565376.2
ENST00000396208.2 |
CCP110
|
centriolar coiled coil protein 110kDa |
| chr19_+_41313017 | 0.22 |
ENST00000595621.1
ENST00000595051.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr16_-_20753114 | 0.22 |
ENST00000396083.2
|
THUMPD1
|
THUMP domain containing 1 |
| chr8_-_26724784 | 0.22 |
ENST00000380573.3
|
ADRA1A
|
adrenoceptor alpha 1A |
| chr11_+_61129456 | 0.22 |
ENST00000278826.6
|
TMEM138
|
transmembrane protein 138 |
| chr15_+_89346699 | 0.22 |
ENST00000558207.1
|
ACAN
|
aggrecan |
| chr8_+_145438870 | 0.21 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B |
| chr17_-_41132010 | 0.21 |
ENST00000409103.1
ENST00000360221.4 |
PTGES3L-AARSD1
|
PTGES3L-AARSD1 readthrough |
| chr4_-_151936865 | 0.21 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr6_+_2246023 | 0.20 |
ENST00000530833.1
ENST00000525811.1 ENST00000534441.1 ENST00000533653.1 ENST00000534468.1 |
GMDS-AS1
|
GMDS antisense RNA 1 (head to head) |
| chr10_-_99531709 | 0.20 |
ENST00000266066.3
|
SFRP5
|
secreted frizzled-related protein 5 |
| chr15_+_89402148 | 0.20 |
ENST00000560601.1
|
ACAN
|
aggrecan |
| chr1_-_109940550 | 0.20 |
ENST00000256637.6
|
SORT1
|
sortilin 1 |
| chr15_+_89346657 | 0.20 |
ENST00000439576.2
|
ACAN
|
aggrecan |
| chr11_+_43702322 | 0.20 |
ENST00000395700.4
|
HSD17B12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr16_-_49315731 | 0.20 |
ENST00000219197.6
|
CBLN1
|
cerebellin 1 precursor |
| chrX_+_22050546 | 0.20 |
ENST00000379374.4
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr8_+_38854368 | 0.19 |
ENST00000466936.1
|
ADAM9
|
ADAM metallopeptidase domain 9 |
| chr6_+_33043703 | 0.19 |
ENST00000418931.2
ENST00000535465.1 |
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr20_-_41818536 | 0.19 |
ENST00000373193.3
ENST00000373198.4 ENST00000373201.1 |
PTPRT
|
protein tyrosine phosphatase, receptor type, T |
| chrX_-_71933888 | 0.19 |
ENST00000373542.4
ENST00000339490.3 ENST00000541944.1 ENST00000373539.3 ENST00000373545.3 |
PHKA1
|
phosphorylase kinase, alpha 1 (muscle) |
| chr20_+_30640004 | 0.19 |
ENST00000520553.1
ENST00000518730.1 ENST00000375852.2 |
HCK
|
hemopoietic cell kinase |
| chr7_-_16844611 | 0.19 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr2_+_46769798 | 0.18 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr13_-_84456527 | 0.18 |
ENST00000377084.2
|
SLITRK1
|
SLIT and NTRK-like family, member 1 |
| chr3_-_156534754 | 0.18 |
ENST00000472943.1
ENST00000473352.1 |
LINC00886
|
long intergenic non-protein coding RNA 886 |
| chr2_-_220435963 | 0.18 |
ENST00000373876.1
ENST00000404537.1 ENST00000603926.1 ENST00000373873.4 ENST00000289656.3 |
OBSL1
|
obscurin-like 1 |
| chr5_+_141016508 | 0.18 |
ENST00000444782.1
ENST00000521367.1 ENST00000297164.3 |
RELL2
|
RELT-like 2 |
| chr2_-_24583314 | 0.18 |
ENST00000443927.1
ENST00000406921.3 ENST00000412011.1 |
ITSN2
|
intersectin 2 |
| chr12_-_55378452 | 0.18 |
ENST00000449076.1
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr9_+_137967268 | 0.17 |
ENST00000371799.4
ENST00000277415.11 |
OLFM1
|
olfactomedin 1 |
| chr5_+_141016969 | 0.17 |
ENST00000518856.1
|
RELL2
|
RELT-like 2 |
| chr19_+_34895289 | 0.17 |
ENST00000246535.3
|
PDCD2L
|
programmed cell death 2-like |
| chr11_+_59522837 | 0.17 |
ENST00000437946.2
|
STX3
|
syntaxin 3 |
| chr15_-_41166414 | 0.17 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr5_-_24645078 | 0.17 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr11_-_76155700 | 0.17 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr14_+_63671577 | 0.17 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr16_-_18937726 | 0.17 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr22_-_39268308 | 0.17 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr14_+_58666824 | 0.16 |
ENST00000254286.4
|
ACTR10
|
actin-related protein 10 homolog (S. cerevisiae) |
| chr8_+_121137333 | 0.16 |
ENST00000309791.4
ENST00000297848.3 ENST00000247781.3 |
COL14A1
|
collagen, type XIV, alpha 1 |
| chr5_+_139027877 | 0.16 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr21_-_40685477 | 0.16 |
ENST00000342449.3
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr6_-_10882174 | 0.16 |
ENST00000379491.4
|
GCM2
|
glial cells missing homolog 2 (Drosophila) |
| chr14_-_101034407 | 0.16 |
ENST00000443071.2
ENST00000557378.1 |
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr2_-_220436248 | 0.16 |
ENST00000265318.4
|
OBSL1
|
obscurin-like 1 |
| chrX_+_53123314 | 0.16 |
ENST00000605526.1
ENST00000604062.1 ENST00000604369.1 ENST00000366185.2 ENST00000604849.1 |
RP11-258C19.5
|
long intergenic non-protein coding RNA 1155 |
| chr16_+_11038403 | 0.16 |
ENST00000409552.3
|
CLEC16A
|
C-type lectin domain family 16, member A |
| chr20_+_60813535 | 0.16 |
ENST00000358053.2
ENST00000313733.3 ENST00000439951.2 |
OSBPL2
|
oxysterol binding protein-like 2 |
| chr20_-_41818373 | 0.16 |
ENST00000373187.1
ENST00000356100.2 ENST00000373184.1 ENST00000373190.1 |
PTPRT
|
protein tyrosine phosphatase, receptor type, T |
| chr7_-_150675372 | 0.16 |
ENST00000262186.5
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr1_-_85930246 | 0.16 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr19_+_496454 | 0.16 |
ENST00000346144.4
ENST00000215637.3 ENST00000382683.4 |
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr11_-_19223523 | 0.15 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr12_+_100867694 | 0.15 |
ENST00000392986.3
ENST00000549996.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr11_+_59522900 | 0.15 |
ENST00000529177.1
|
STX3
|
syntaxin 3 |
| chr1_-_197744322 | 0.15 |
ENST00000235453.4
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr17_+_79989500 | 0.15 |
ENST00000306897.4
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr17_-_8151353 | 0.15 |
ENST00000315684.8
|
CTC1
|
CTS telomere maintenance complex component 1 |
| chr3_-_67705006 | 0.15 |
ENST00000492795.1
ENST00000493112.1 ENST00000307227.5 |
SUCLG2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr17_+_79679299 | 0.15 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr6_+_2245982 | 0.15 |
ENST00000530346.1
ENST00000524770.1 ENST00000532124.1 ENST00000531092.1 ENST00000456943.2 ENST00000529893.1 |
GMDS-AS1
|
GMDS antisense RNA 1 (head to head) |
| chr20_+_30639991 | 0.14 |
ENST00000534862.1
ENST00000538448.1 ENST00000375862.2 |
HCK
|
hemopoietic cell kinase |
| chr14_-_103523745 | 0.14 |
ENST00000361246.2
|
CDC42BPB
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr1_-_33168336 | 0.14 |
ENST00000373484.3
|
SYNC
|
syncoilin, intermediate filament protein |
| chr16_+_15528332 | 0.14 |
ENST00000566490.1
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr6_+_20403997 | 0.14 |
ENST00000535432.1
|
E2F3
|
E2F transcription factor 3 |
| chr6_-_89927151 | 0.14 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr1_-_197744390 | 0.14 |
ENST00000367396.3
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr13_-_38443860 | 0.14 |
ENST00000426868.2
ENST00000379681.3 ENST00000338947.5 ENST00000355779.2 ENST00000358477.2 ENST00000379673.2 |
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr9_-_26892765 | 0.14 |
ENST00000520187.1
ENST00000333916.5 |
CAAP1
|
caspase activity and apoptosis inhibitor 1 |
| chr17_+_79679369 | 0.14 |
ENST00000350690.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr3_+_49711777 | 0.14 |
ENST00000442186.1
ENST00000438011.1 ENST00000457042.1 |
APEH
|
acylaminoacyl-peptide hydrolase |
| chr16_+_28996572 | 0.13 |
ENST00000360872.5
ENST00000566177.1 ENST00000354453.4 |
LAT
|
linker for activation of T cells |
| chr4_+_86396265 | 0.13 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr4_-_151936416 | 0.13 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr12_+_7060432 | 0.13 |
ENST00000318974.9
ENST00000456013.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr6_-_8102714 | 0.13 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr19_-_10697895 | 0.13 |
ENST00000591240.1
ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr17_+_42836329 | 0.13 |
ENST00000200557.6
|
ADAM11
|
ADAM metallopeptidase domain 11 |
| chr2_-_175462934 | 0.13 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr1_-_54405773 | 0.13 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr16_+_29984962 | 0.13 |
ENST00000308893.4
|
TAOK2
|
TAO kinase 2 |
| chr19_+_38085768 | 0.13 |
ENST00000316433.4
ENST00000590588.1 ENST00000586134.1 ENST00000586792.1 |
ZNF540
|
zinc finger protein 540 |
| chr14_-_51561784 | 0.13 |
ENST00000360392.4
|
TRIM9
|
tripartite motif containing 9 |
| chr20_+_49348081 | 0.13 |
ENST00000371610.2
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr19_-_18385221 | 0.13 |
ENST00000595654.2
ENST00000593659.1 ENST00000599528.1 |
KIAA1683
|
KIAA1683 |
| chr22_-_24093267 | 0.13 |
ENST00000341976.3
|
ZNF70
|
zinc finger protein 70 |
| chr1_+_202431859 | 0.12 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr14_+_105781048 | 0.12 |
ENST00000458164.2
ENST00000447393.1 |
PACS2
|
phosphofurin acidic cluster sorting protein 2 |
| chr16_+_11038345 | 0.12 |
ENST00000409790.1
|
CLEC16A
|
C-type lectin domain family 16, member A |
| chr10_-_7708918 | 0.12 |
ENST00000256861.6
ENST00000397146.2 ENST00000446830.2 ENST00000397145.2 |
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr4_-_54457783 | 0.12 |
ENST00000263925.7
ENST00000512247.1 |
LNX1
|
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| chrX_-_153285251 | 0.12 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr17_-_47755338 | 0.12 |
ENST00000508805.1
ENST00000515508.2 ENST00000451526.2 ENST00000507970.1 |
SPOP
|
speckle-type POZ protein |
| chr9_-_130616915 | 0.11 |
ENST00000344849.3
|
ENG
|
endoglin |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.4 | 1.3 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) regulation of lactation(GO:1903487) |
| 0.2 | 3.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.2 | 3.8 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 0.6 | GO:0034125 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 0.2 | 1.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.2 | 1.7 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.7 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 1.0 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.4 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.3 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.4 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.3 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.1 | 0.3 | GO:1902356 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 0.5 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.3 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.1 | 1.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 0.3 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 1.8 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 | 0.2 | GO:0003099 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.1 | 0.2 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.1 | 2.1 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 1.9 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.1 | 0.5 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.4 | GO:0033504 | floor plate development(GO:0033504) |
| 0.1 | 0.2 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.3 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.2 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.0 | 0.3 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.3 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.4 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.2 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:0034241 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.3 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.2 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.0 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.3 | GO:0021894 | regulation of neuron maturation(GO:0014041) cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0090027 | negative regulation of SMAD protein import into nucleus(GO:0060392) negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.0 | 0.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.6 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.3 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.5 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.4 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.8 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.3 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 0.1 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.6 | GO:0090662 | ATP hydrolysis coupled proton transport(GO:0015991) phagosome acidification(GO:0090383) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.1 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 0.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.5 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.5 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.4 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.2 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 | 0.2 | GO:1900038 | negative regulation of vascular permeability(GO:0043116) negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.3 | 1.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 0.7 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 0.5 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 1.4 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 0.3 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 2.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.2 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.3 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.3 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.6 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0032279 | axonemal microtubule(GO:0005879) asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.3 | 1.6 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.3 | 3.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.2 | 0.6 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 0.7 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.2 | 2.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 1.0 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 0.3 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.2 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.2 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.3 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.6 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.3 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0055131 | phosphorelay sensor kinase activity(GO:0000155) C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.0 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 3.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |