Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
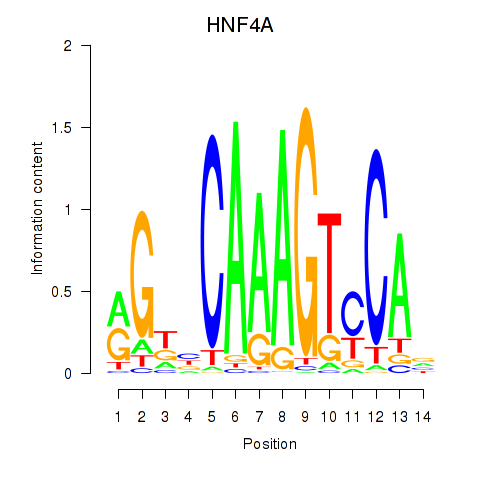
Results for HNF4A
Z-value: 0.51
Transcription factors associated with HNF4A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF4A
|
ENSG00000101076.12 | hepatocyte nuclear factor 4 alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HNF4A | hg19_v2_chr20_+_42984330_42984445 | -0.15 | 4.2e-01 | Click! |
Activity profile of HNF4A motif
Sorted Z-values of HNF4A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_61513146 | 1.78 |
ENST00000430431.1
|
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr20_-_7921090 | 1.61 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr10_-_61513201 | 1.51 |
ENST00000414264.1
ENST00000594536.1 |
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr19_+_41594377 | 1.29 |
ENST00000330436.3
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr2_-_28113217 | 1.18 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr11_+_60467047 | 1.03 |
ENST00000300226.2
|
MS4A8
|
membrane-spanning 4-domains, subfamily A, member 8 |
| chr12_+_56075330 | 1.03 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr2_-_159313214 | 0.88 |
ENST00000409889.1
ENST00000283233.5 ENST00000536771.1 |
CCDC148
|
coiled-coil domain containing 148 |
| chr6_+_116937636 | 0.82 |
ENST00000368581.4
ENST00000229554.5 ENST00000368580.4 |
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr10_-_17171817 | 0.70 |
ENST00000377833.4
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr19_+_4639514 | 0.69 |
ENST00000327473.4
|
TNFAIP8L1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr16_-_16317321 | 0.65 |
ENST00000205557.7
ENST00000575728.1 |
ABCC6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
| chr10_+_96522361 | 0.61 |
ENST00000371321.3
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19 |
| chr5_-_16509101 | 0.56 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr1_-_177939348 | 0.56 |
ENST00000464631.2
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr17_+_7591639 | 0.51 |
ENST00000396463.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr17_+_7591747 | 0.50 |
ENST00000534050.1
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr11_+_63137251 | 0.50 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chr14_+_23352374 | 0.50 |
ENST00000267396.4
ENST00000536884.1 |
REM2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr10_+_96698406 | 0.48 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr2_+_241544834 | 0.48 |
ENST00000319838.5
ENST00000403859.1 ENST00000438013.2 |
GPR35
|
G protein-coupled receptor 35 |
| chr1_-_43751230 | 0.40 |
ENST00000523677.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr7_+_120628731 | 0.39 |
ENST00000310396.5
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr2_+_74648848 | 0.39 |
ENST00000409791.1
ENST00000426787.1 ENST00000348227.4 |
WDR54
|
WD repeat domain 54 |
| chr10_+_96443204 | 0.33 |
ENST00000339022.5
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr15_-_41120896 | 0.33 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr14_-_76447336 | 0.31 |
ENST00000556285.1
|
TGFB3
|
transforming growth factor, beta 3 |
| chr17_-_38256973 | 0.30 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr16_+_19429018 | 0.30 |
ENST00000542583.2
|
TMC5
|
transmembrane channel-like 5 |
| chr1_-_197036364 | 0.30 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr11_+_118868830 | 0.30 |
ENST00000334418.1
|
CCDC84
|
coiled-coil domain containing 84 |
| chr10_+_96443378 | 0.30 |
ENST00000285979.6
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr1_-_155270770 | 0.29 |
ENST00000392414.3
|
PKLR
|
pyruvate kinase, liver and RBC |
| chr19_+_45449301 | 0.28 |
ENST00000591597.1
|
APOC2
|
apolipoprotein C-II |
| chr14_-_76447494 | 0.28 |
ENST00000238682.3
|
TGFB3
|
transforming growth factor, beta 3 |
| chr5_+_79950463 | 0.28 |
ENST00000265081.6
|
MSH3
|
mutS homolog 3 |
| chr14_+_100485712 | 0.28 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr11_-_123756334 | 0.27 |
ENST00000528595.1
ENST00000375026.2 |
TMEM225
|
transmembrane protein 225 |
| chrX_+_38211777 | 0.26 |
ENST00000039007.4
|
OTC
|
ornithine carbamoyltransferase |
| chr1_+_196857144 | 0.26 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr2_+_219433281 | 0.25 |
ENST00000273064.6
ENST00000509807.2 ENST00000542068.1 |
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr11_+_1856034 | 0.25 |
ENST00000341958.3
|
SYT8
|
synaptotagmin VIII |
| chr20_-_44485835 | 0.25 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr12_-_39837192 | 0.24 |
ENST00000361961.3
ENST00000395670.3 |
KIF21A
|
kinesin family member 21A |
| chr22_-_31063782 | 0.24 |
ENST00000404885.1
ENST00000403268.1 ENST00000407308.1 ENST00000342474.4 ENST00000334679.3 |
DUSP18
|
dual specificity phosphatase 18 |
| chr2_-_219433014 | 0.24 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr11_+_1855645 | 0.24 |
ENST00000381968.3
ENST00000381978.3 |
SYT8
|
synaptotagmin VIII |
| chr3_-_194072019 | 0.23 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr9_-_116065551 | 0.23 |
ENST00000297894.5
|
RNF183
|
ring finger protein 183 |
| chr17_-_7082668 | 0.21 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr16_-_3350614 | 0.21 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr19_-_10687907 | 0.21 |
ENST00000589348.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr1_+_78511586 | 0.21 |
ENST00000370759.3
|
GIPC2
|
GIPC PDZ domain containing family, member 2 |
| chr11_-_10315741 | 0.20 |
ENST00000256190.8
|
SBF2
|
SET binding factor 2 |
| chr3_-_158390282 | 0.20 |
ENST00000264265.3
|
LXN
|
latexin |
| chr5_-_35048047 | 0.19 |
ENST00000231420.6
|
AGXT2
|
alanine--glyoxylate aminotransferase 2 |
| chr11_-_64660916 | 0.19 |
ENST00000413053.1
|
MIR194-2
|
microRNA 194-2 |
| chr19_-_48867291 | 0.19 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr3_+_123813509 | 0.19 |
ENST00000460856.1
ENST00000240874.3 |
KALRN
|
kalirin, RhoGEF kinase |
| chr11_-_32456891 | 0.19 |
ENST00000452863.3
|
WT1
|
Wilms tumor 1 |
| chr1_+_6508100 | 0.18 |
ENST00000461727.1
|
ESPN
|
espin |
| chr20_+_57875457 | 0.18 |
ENST00000337938.2
ENST00000311585.7 ENST00000371028.2 |
EDN3
|
endothelin 3 |
| chr6_+_46761118 | 0.18 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr17_+_4675175 | 0.18 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr1_+_43855560 | 0.17 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr6_-_86099898 | 0.17 |
ENST00000455071.1
|
RP11-30P6.6
|
RP11-30P6.6 |
| chr9_+_27109133 | 0.17 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr10_-_116444371 | 0.16 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr6_+_30131318 | 0.16 |
ENST00000376688.1
|
TRIM15
|
tripartite motif containing 15 |
| chr1_+_241695424 | 0.16 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr4_-_120225600 | 0.15 |
ENST00000399075.4
|
C4orf3
|
chromosome 4 open reading frame 3 |
| chr2_+_159313452 | 0.15 |
ENST00000389757.3
ENST00000389759.3 |
PKP4
|
plakophilin 4 |
| chr2_-_62733476 | 0.15 |
ENST00000335390.5
|
TMEM17
|
transmembrane protein 17 |
| chr6_-_31926629 | 0.15 |
ENST00000375425.5
ENST00000426722.1 ENST00000441998.1 ENST00000444811.2 ENST00000375429.3 |
NELFE
|
negative elongation factor complex member E |
| chr16_+_24857162 | 0.15 |
ENST00000347898.3
|
SLC5A11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr16_-_88752889 | 0.15 |
ENST00000332281.5
|
SNAI3
|
snail family zinc finger 3 |
| chr13_+_50656307 | 0.14 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr1_-_177939041 | 0.14 |
ENST00000308284.6
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr1_-_60392452 | 0.14 |
ENST00000371204.3
|
CYP2J2
|
cytochrome P450, family 2, subfamily J, polypeptide 2 |
| chr16_+_24857552 | 0.14 |
ENST00000568579.1
ENST00000567758.1 ENST00000569071.1 ENST00000539472.1 |
SLC5A11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr1_+_180601139 | 0.14 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr19_-_48867171 | 0.14 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr10_-_96829246 | 0.14 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr16_+_24857309 | 0.14 |
ENST00000565769.1
ENST00000449109.2 ENST00000424767.2 ENST00000545376.1 ENST00000569520.1 |
SLC5A11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr20_+_56136136 | 0.14 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr14_+_77648167 | 0.13 |
ENST00000554346.1
ENST00000298351.4 |
TMEM63C
|
transmembrane protein 63C |
| chr22_+_25003606 | 0.13 |
ENST00000432867.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr17_+_27369918 | 0.13 |
ENST00000323372.4
|
PIPOX
|
pipecolic acid oxidase |
| chr17_+_41857793 | 0.13 |
ENST00000449302.3
|
C17orf105
|
chromosome 17 open reading frame 105 |
| chr11_-_73720276 | 0.13 |
ENST00000348534.4
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr3_+_63897605 | 0.13 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr6_+_30130969 | 0.12 |
ENST00000376694.4
|
TRIM15
|
tripartite motif containing 15 |
| chr5_+_133842243 | 0.12 |
ENST00000515627.2
|
AC005355.2
|
AC005355.2 |
| chr6_+_31982539 | 0.12 |
ENST00000435363.2
ENST00000425700.2 |
C4B
|
complement component 4B (Chido blood group) |
| chrX_+_118108571 | 0.11 |
ENST00000304778.7
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr19_+_46806856 | 0.11 |
ENST00000300862.3
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit |
| chr19_+_45449228 | 0.11 |
ENST00000252490.4
|
APOC2
|
apolipoprotein C-II |
| chr2_+_219433588 | 0.11 |
ENST00000295701.5
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr3_+_108541545 | 0.11 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr9_+_139839686 | 0.11 |
ENST00000371634.2
|
C8G
|
complement component 8, gamma polypeptide |
| chr7_-_37956409 | 0.11 |
ENST00000436072.2
|
SFRP4
|
secreted frizzled-related protein 4 |
| chr3_+_108541608 | 0.10 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chrX_+_66764375 | 0.10 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr1_-_15911510 | 0.10 |
ENST00000375826.3
|
AGMAT
|
agmatine ureohydrolase (agmatinase) |
| chr15_-_73925651 | 0.10 |
ENST00000545878.1
ENST00000287226.8 ENST00000345330.4 |
NPTN
|
neuroplastin |
| chr20_-_3644046 | 0.10 |
ENST00000290417.2
ENST00000319242.3 |
GFRA4
|
GDNF family receptor alpha 4 |
| chr12_+_109273806 | 0.10 |
ENST00000228476.3
ENST00000547768.1 |
DAO
|
D-amino-acid oxidase |
| chr1_+_241695670 | 0.10 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr17_+_77704681 | 0.10 |
ENST00000328313.5
|
ENPP7
|
ectonucleotide pyrophosphatase/phosphodiesterase 7 |
| chr20_+_57875658 | 0.10 |
ENST00000371025.3
|
EDN3
|
endothelin 3 |
| chr9_+_139839711 | 0.09 |
ENST00000224181.3
|
C8G
|
complement component 8, gamma polypeptide |
| chr2_-_21266935 | 0.09 |
ENST00000233242.1
|
APOB
|
apolipoprotein B |
| chr6_-_29343068 | 0.09 |
ENST00000396806.3
|
OR12D3
|
olfactory receptor, family 12, subfamily D, member 3 |
| chr12_+_56114189 | 0.09 |
ENST00000548082.1
|
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr11_-_61658853 | 0.09 |
ENST00000525588.1
ENST00000540820.1 |
FADS3
|
fatty acid desaturase 3 |
| chr8_+_104831554 | 0.09 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr2_-_11810284 | 0.09 |
ENST00000306928.5
|
NTSR2
|
neurotensin receptor 2 |
| chr3_+_124303539 | 0.09 |
ENST00000428018.2
|
KALRN
|
kalirin, RhoGEF kinase |
| chr13_+_76334498 | 0.08 |
ENST00000534657.1
|
LMO7
|
LIM domain 7 |
| chr6_+_31949801 | 0.08 |
ENST00000428956.2
ENST00000498271.1 |
C4A
|
complement component 4A (Rodgers blood group) |
| chr17_+_4853442 | 0.08 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr19_-_10687983 | 0.08 |
ENST00000587069.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr15_+_81489213 | 0.08 |
ENST00000559383.1
ENST00000394660.2 |
IL16
|
interleukin 16 |
| chr3_-_187009798 | 0.08 |
ENST00000337774.5
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr17_+_44039704 | 0.08 |
ENST00000420682.2
ENST00000415613.2 ENST00000571987.1 ENST00000574436.1 ENST00000431008.3 |
MAPT
|
microtubule-associated protein tau |
| chr22_+_25003568 | 0.08 |
ENST00000447416.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr12_-_46384334 | 0.08 |
ENST00000369367.3
ENST00000266589.6 ENST00000395453.2 ENST00000395454.2 |
SCAF11
|
SR-related CTD-associated factor 11 |
| chrX_+_65382433 | 0.08 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr11_+_64073699 | 0.07 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr12_+_56114151 | 0.07 |
ENST00000547072.1
ENST00000552930.1 ENST00000257895.5 |
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr19_-_12886327 | 0.07 |
ENST00000397668.3
ENST00000587178.1 ENST00000264827.5 |
HOOK2
|
hook microtubule-tethering protein 2 |
| chr3_-_124653579 | 0.07 |
ENST00000478191.1
ENST00000311075.3 |
MUC13
|
mucin 13, cell surface associated |
| chr3_+_63898275 | 0.07 |
ENST00000538065.1
|
ATXN7
|
ataxin 7 |
| chr3_-_49851313 | 0.07 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr10_+_70320413 | 0.07 |
ENST00000373644.4
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr6_+_43028182 | 0.07 |
ENST00000394058.1
|
KLC4
|
kinesin light chain 4 |
| chr3_+_123813543 | 0.07 |
ENST00000360013.3
|
KALRN
|
kalirin, RhoGEF kinase |
| chr19_-_10687948 | 0.06 |
ENST00000592285.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr17_+_61554413 | 0.06 |
ENST00000538928.1
ENST00000290866.4 ENST00000428043.1 |
ACE
|
angiotensin I converting enzyme |
| chrX_+_118108601 | 0.06 |
ENST00000371628.3
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr5_-_32444828 | 0.06 |
ENST00000265069.8
|
ZFR
|
zinc finger RNA binding protein |
| chr1_+_100435315 | 0.05 |
ENST00000370155.3
ENST00000465289.1 |
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr11_-_45939374 | 0.05 |
ENST00000533151.1
ENST00000241041.3 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr7_+_45928079 | 0.05 |
ENST00000468955.1
|
IGFBP1
|
insulin-like growth factor binding protein 1 |
| chr14_+_23790655 | 0.05 |
ENST00000397276.2
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr20_+_57875758 | 0.05 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr15_+_22382382 | 0.05 |
ENST00000328795.4
|
OR4N4
|
olfactory receptor, family 4, subfamily N, member 4 |
| chr1_+_94883991 | 0.05 |
ENST00000370214.4
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr3_+_46538981 | 0.05 |
ENST00000296142.3
|
RTP3
|
receptor (chemosensory) transporter protein 3 |
| chr5_+_72469014 | 0.05 |
ENST00000296776.5
|
TMEM174
|
transmembrane protein 174 |
| chr19_-_7698599 | 0.05 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr10_-_103815874 | 0.05 |
ENST00000370033.4
ENST00000311122.5 |
C10orf76
|
chromosome 10 open reading frame 76 |
| chr12_+_67663056 | 0.05 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr18_+_54318616 | 0.05 |
ENST00000254442.3
|
WDR7
|
WD repeat domain 7 |
| chr14_+_23790690 | 0.04 |
ENST00000556821.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr13_+_52586517 | 0.04 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr9_-_27005686 | 0.04 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr19_+_39881951 | 0.04 |
ENST00000315588.5
ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29
|
mediator complex subunit 29 |
| chr7_+_45927956 | 0.04 |
ENST00000275525.3
ENST00000457280.1 |
IGFBP1
|
insulin-like growth factor binding protein 1 |
| chr17_+_41052808 | 0.04 |
ENST00000592383.1
ENST00000253801.2 ENST00000585489.1 |
G6PC
|
glucose-6-phosphatase, catalytic subunit |
| chr18_+_54318566 | 0.04 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr9_-_15307200 | 0.04 |
ENST00000506891.1
ENST00000541445.1 ENST00000512701.2 ENST00000380850.4 ENST00000297615.5 ENST00000355694.2 |
TTC39B
|
tetratricopeptide repeat domain 39B |
| chr19_+_45449266 | 0.04 |
ENST00000592257.1
|
APOC2
|
apolipoprotein C-II |
| chr1_-_43855479 | 0.04 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr15_+_50716576 | 0.04 |
ENST00000560297.1
ENST00000307179.4 ENST00000396444.3 ENST00000433963.1 ENST00000425032.3 |
USP8
|
ubiquitin specific peptidase 8 |
| chr3_-_141868357 | 0.04 |
ENST00000489671.1
ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr20_+_36661910 | 0.04 |
ENST00000373433.4
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr6_+_143999072 | 0.04 |
ENST00000440869.2
ENST00000367582.3 ENST00000451827.2 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr6_-_149806105 | 0.04 |
ENST00000389942.5
ENST00000416573.2 ENST00000542614.1 ENST00000409806.3 |
ZC3H12D
|
zinc finger CCCH-type containing 12D |
| chr15_-_100273544 | 0.04 |
ENST00000409796.1
ENST00000545021.1 ENST00000344791.2 ENST00000332728.4 ENST00000450512.1 |
LYSMD4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr16_-_87970122 | 0.03 |
ENST00000309893.2
|
CA5A
|
carbonic anhydrase VA, mitochondrial |
| chr17_-_2614927 | 0.03 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr13_+_76334567 | 0.03 |
ENST00000321797.8
|
LMO7
|
LIM domain 7 |
| chr12_-_7125770 | 0.03 |
ENST00000261407.4
|
LPCAT3
|
lysophosphatidylcholine acyltransferase 3 |
| chr1_-_161208013 | 0.03 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr19_-_39881777 | 0.03 |
ENST00000595564.1
ENST00000221265.3 |
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr1_-_41131326 | 0.03 |
ENST00000372684.3
|
RIMS3
|
regulating synaptic membrane exocytosis 3 |
| chr2_+_10442993 | 0.03 |
ENST00000423674.1
ENST00000307845.3 |
HPCAL1
|
hippocalcin-like 1 |
| chr20_+_44486246 | 0.03 |
ENST00000255152.2
ENST00000454862.2 |
ZSWIM3
|
zinc finger, SWIM-type containing 3 |
| chr13_+_76334795 | 0.03 |
ENST00000526202.1
ENST00000465261.2 |
LMO7
|
LIM domain 7 |
| chr6_-_33385854 | 0.03 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr6_-_33385823 | 0.03 |
ENST00000494751.1
ENST00000374496.3 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr6_+_80341000 | 0.03 |
ENST00000369838.4
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr15_-_101142401 | 0.03 |
ENST00000314742.8
|
LINS
|
lines homolog (Drosophila) |
| chr6_-_33385655 | 0.03 |
ENST00000440279.3
ENST00000607266.1 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr11_-_73720122 | 0.02 |
ENST00000426995.2
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr5_-_42811986 | 0.02 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr2_-_219134822 | 0.02 |
ENST00000444053.1
ENST00000248450.4 |
AAMP
|
angio-associated, migratory cell protein |
| chr12_-_51422017 | 0.02 |
ENST00000394904.3
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr14_+_64680854 | 0.02 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr10_+_52152766 | 0.02 |
ENST00000596442.1
|
AC069547.2
|
Uncharacterized protein |
| chr1_-_161207953 | 0.02 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr10_+_104404218 | 0.02 |
ENST00000302424.7
|
TRIM8
|
tripartite motif containing 8 |
| chr7_+_192969 | 0.02 |
ENST00000313766.5
|
FAM20C
|
family with sequence similarity 20, member C |
| chr12_-_21757774 | 0.02 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
| chr6_+_31926857 | 0.02 |
ENST00000375394.2
ENST00000544581.1 |
SKIV2L
|
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr1_-_167059830 | 0.02 |
ENST00000367868.3
|
GPA33
|
glycoprotein A33 (transmembrane) |
| chr2_-_179914760 | 0.02 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr8_+_96037205 | 0.02 |
ENST00000396124.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr12_+_121163538 | 0.02 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
Network of associatons between targets according to the STRING database.
First level regulatory network of HNF4A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.2 | 1.2 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.2 | 1.6 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.2 | 0.6 | GO:0042704 | detection of oxygen(GO:0003032) uterine wall breakdown(GO:0042704) |
| 0.2 | 1.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.2 | 1.2 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.1 | 0.5 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.4 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.3 | GO:0070859 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 0.3 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 0.3 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.1 | 0.5 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.4 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 0.7 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.2 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 0.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.9 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.3 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.2 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.3 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.3 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) response to selenium ion(GO:0010269) |
| 0.0 | 0.0 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.8 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.6 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.3 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.7 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.5 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 2.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 1.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.2 | 3.3 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.3 | GO:0000406 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.2 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 1.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 1.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.4 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.0 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 1.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.0 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |