Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
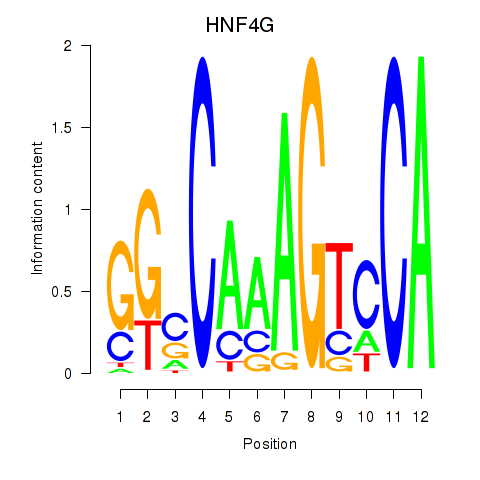
Results for HNF4G
Z-value: 1.28
Transcription factors associated with HNF4G
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF4G
|
ENSG00000164749.7 | hepatocyte nuclear factor 4 gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HNF4G | hg19_v2_chr8_+_76452097_76452126 | 0.71 | 9.5e-06 | Click! |
Activity profile of HNF4G motif
Sorted Z-values of HNF4G motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_41356347 | 14.45 |
ENST00000301141.5
|
CYP2A6
|
cytochrome P450, family 2, subfamily A, polypeptide 6 |
| chr4_-_7044657 | 7.25 |
ENST00000310085.4
|
CCDC96
|
coiled-coil domain containing 96 |
| chr19_-_41388657 | 6.70 |
ENST00000301146.4
ENST00000291764.3 |
CYP2A7
|
cytochrome P450, family 2, subfamily A, polypeptide 7 |
| chr21_+_42688657 | 6.66 |
ENST00000357985.2
|
FAM3B
|
family with sequence similarity 3, member B |
| chr5_+_94727048 | 6.59 |
ENST00000283357.5
|
FAM81B
|
family with sequence similarity 81, member B |
| chr4_-_16077741 | 6.36 |
ENST00000447510.2
ENST00000540805.1 ENST00000539194.1 |
PROM1
|
prominin 1 |
| chr9_-_112970436 | 5.91 |
ENST00000400613.4
|
C9orf152
|
chromosome 9 open reading frame 152 |
| chr19_+_41594377 | 5.71 |
ENST00000330436.3
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr14_+_105952648 | 5.67 |
ENST00000330233.7
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr4_+_165675197 | 5.61 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr3_-_19975665 | 5.04 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chr16_-_776431 | 4.78 |
ENST00000293889.6
|
CCDC78
|
coiled-coil domain containing 78 |
| chr16_+_84209539 | 4.38 |
ENST00000569735.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr2_-_99771373 | 4.37 |
ENST00000393483.3
|
TSGA10
|
testis specific, 10 |
| chr19_+_4639514 | 4.16 |
ENST00000327473.4
|
TNFAIP8L1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr3_+_100354442 | 4.14 |
ENST00000475887.1
|
GPR128
|
G protein-coupled receptor 128 |
| chr4_+_165675269 | 4.10 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr1_-_183622442 | 3.66 |
ENST00000308641.4
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr14_-_106926724 | 3.61 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr17_+_68165657 | 3.51 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr1_+_236686875 | 3.48 |
ENST00000366584.4
|
LGALS8
|
lectin, galactoside-binding, soluble, 8 |
| chr1_+_18807424 | 3.39 |
ENST00000400664.1
|
KLHDC7A
|
kelch domain containing 7A |
| chr2_-_220110111 | 3.24 |
ENST00000428427.1
ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L
|
galactosidase, beta 1-like |
| chr2_-_220110187 | 3.23 |
ENST00000295759.7
ENST00000392089.2 |
GLB1L
|
galactosidase, beta 1-like |
| chr16_+_57406368 | 3.15 |
ENST00000006053.6
ENST00000563383.1 |
CX3CL1
|
chemokine (C-X3-C motif) ligand 1 |
| chr1_+_236686717 | 3.11 |
ENST00000341872.6
ENST00000450372.2 |
LGALS8
|
lectin, galactoside-binding, soluble, 8 |
| chr3_-_52488048 | 3.06 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chr2_-_238499303 | 3.02 |
ENST00000409576.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr6_+_112408768 | 2.97 |
ENST00000368656.2
ENST00000604268.1 |
FAM229B
|
family with sequence similarity 229, member B |
| chr19_-_6720686 | 2.88 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr10_+_96443204 | 2.86 |
ENST00000339022.5
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr10_+_96443378 | 2.81 |
ENST00000285979.6
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr19_-_9003586 | 2.79 |
ENST00000380951.5
|
MUC16
|
mucin 16, cell surface associated |
| chr16_+_811073 | 2.74 |
ENST00000382862.3
ENST00000563651.1 |
MSLN
|
mesothelin |
| chr16_+_2880157 | 2.72 |
ENST00000382280.3
|
ZG16B
|
zymogen granule protein 16B |
| chr3_-_46506358 | 2.66 |
ENST00000417439.1
ENST00000431944.1 |
LTF
|
lactotransferrin |
| chr8_+_27348649 | 2.65 |
ENST00000521780.1
ENST00000380476.3 ENST00000518379.1 ENST00000521684.1 |
EPHX2
|
epoxide hydrolase 2, cytoplasmic |
| chr12_-_122107549 | 2.62 |
ENST00000355329.3
|
MORN3
|
MORN repeat containing 3 |
| chr3_+_100328433 | 2.62 |
ENST00000273352.3
|
GPR128
|
G protein-coupled receptor 128 |
| chr11_+_63137251 | 2.54 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chr9_-_130639997 | 2.54 |
ENST00000373176.1
|
AK1
|
adenylate kinase 1 |
| chr8_+_27348626 | 2.53 |
ENST00000517536.1
|
EPHX2
|
epoxide hydrolase 2, cytoplasmic |
| chr14_-_94854926 | 2.47 |
ENST00000402629.1
ENST00000556091.1 ENST00000554720.1 |
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr19_-_55686399 | 2.45 |
ENST00000587067.1
|
SYT5
|
synaptotagmin V |
| chr17_+_62075703 | 2.41 |
ENST00000577953.1
ENST00000582540.1 ENST00000579184.1 ENST00000425164.3 ENST00000412177.1 ENST00000539996.1 ENST00000583891.1 ENST00000580752.1 |
C17orf72
|
chromosome 17 open reading frame 72 |
| chr2_-_238499131 | 2.37 |
ENST00000538644.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr11_-_111175739 | 2.37 |
ENST00000532918.1
|
COLCA1
|
colorectal cancer associated 1 |
| chr1_+_246887349 | 2.36 |
ENST00000366510.3
|
SCCPDH
|
saccharopine dehydrogenase (putative) |
| chr16_+_2880254 | 2.30 |
ENST00000570670.1
|
ZG16B
|
zymogen granule protein 16B |
| chr11_-_118023490 | 2.28 |
ENST00000324727.4
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr14_+_52781079 | 2.28 |
ENST00000245457.5
|
PTGER2
|
prostaglandin E receptor 2 (subtype EP2), 53kDa |
| chr2_-_238499725 | 2.27 |
ENST00000264601.3
|
RAB17
|
RAB17, member RAS oncogene family |
| chr15_-_78526855 | 2.21 |
ENST00000541759.1
ENST00000558130.1 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr15_-_68497657 | 2.18 |
ENST00000448060.2
ENST00000467889.1 |
CALML4
|
calmodulin-like 4 |
| chr3_-_46506563 | 2.15 |
ENST00000231751.4
|
LTF
|
lactotransferrin |
| chr17_+_7608511 | 2.14 |
ENST00000226091.2
|
EFNB3
|
ephrin-B3 |
| chr14_-_31889782 | 2.08 |
ENST00000543095.2
|
HEATR5A
|
HEAT repeat containing 5A |
| chr2_-_238499337 | 2.06 |
ENST00000411462.1
ENST00000409822.1 |
RAB17
|
RAB17, member RAS oncogene family |
| chr14_-_92333873 | 2.06 |
ENST00000435962.2
|
TC2N
|
tandem C2 domains, nuclear |
| chr2_+_98330009 | 2.00 |
ENST00000264972.5
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa |
| chr18_-_53253112 | 1.96 |
ENST00000568673.1
ENST00000562847.1 ENST00000568147.1 |
TCF4
|
transcription factor 4 |
| chr5_+_35617940 | 1.95 |
ENST00000282469.6
ENST00000509059.1 ENST00000356031.3 ENST00000510777.1 |
SPEF2
|
sperm flagellar 2 |
| chr10_-_100027943 | 1.93 |
ENST00000260702.3
|
LOXL4
|
lysyl oxidase-like 4 |
| chr11_+_113185251 | 1.92 |
ENST00000529221.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr12_+_119772502 | 1.91 |
ENST00000536742.1
ENST00000327554.2 |
CCDC60
|
coiled-coil domain containing 60 |
| chr18_-_53253323 | 1.90 |
ENST00000540999.1
ENST00000563888.2 |
TCF4
|
transcription factor 4 |
| chr11_-_5271122 | 1.85 |
ENST00000330597.3
|
HBG1
|
hemoglobin, gamma A |
| chr4_+_3465027 | 1.83 |
ENST00000389653.2
ENST00000507039.1 ENST00000340083.5 |
DOK7
|
docking protein 7 |
| chr12_+_121163538 | 1.81 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chrX_-_48693955 | 1.71 |
ENST00000218230.5
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr16_+_25228242 | 1.69 |
ENST00000219660.5
|
AQP8
|
aquaporin 8 |
| chr2_-_222436988 | 1.68 |
ENST00000409854.1
ENST00000281821.2 ENST00000392071.4 ENST00000443796.1 |
EPHA4
|
EPH receptor A4 |
| chr7_-_138482849 | 1.67 |
ENST00000353492.4
|
ATP6V0A4
|
ATPase, H+ transporting, lysosomal V0 subunit a4 |
| chr7_+_138482695 | 1.64 |
ENST00000422794.2
ENST00000397602.3 ENST00000442682.2 ENST00000458494.1 ENST00000413208.1 |
TMEM213
|
transmembrane protein 213 |
| chr11_+_1856034 | 1.64 |
ENST00000341958.3
|
SYT8
|
synaptotagmin VIII |
| chr16_-_19896220 | 1.63 |
ENST00000562469.1
ENST00000300571.2 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr2_-_31030277 | 1.59 |
ENST00000534090.2
ENST00000295055.8 |
CAPN13
|
calpain 13 |
| chr22_+_29279552 | 1.59 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr12_+_20963647 | 1.59 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chrX_-_151999269 | 1.56 |
ENST00000370277.3
|
CETN2
|
centrin, EF-hand protein, 2 |
| chr11_-_63330842 | 1.54 |
ENST00000255695.1
|
HRASLS2
|
HRAS-like suppressor 2 |
| chr2_+_230787201 | 1.54 |
ENST00000283946.3
|
FBXO36
|
F-box protein 36 |
| chr2_+_230787213 | 1.54 |
ENST00000409992.1
|
FBXO36
|
F-box protein 36 |
| chr12_+_20963632 | 1.52 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr10_-_69597915 | 1.51 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr19_+_44645731 | 1.51 |
ENST00000426739.2
|
ZNF234
|
zinc finger protein 234 |
| chr1_+_164528866 | 1.50 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr17_-_63556414 | 1.45 |
ENST00000585045.1
|
AXIN2
|
axin 2 |
| chr16_-_3350614 | 1.41 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr16_+_19179549 | 1.39 |
ENST00000355377.2
ENST00000568115.1 |
SYT17
|
synaptotagmin XVII |
| chr16_+_83932684 | 1.36 |
ENST00000262430.4
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr3_-_47324079 | 1.35 |
ENST00000352910.4
|
KIF9
|
kinesin family member 9 |
| chr1_+_236687173 | 1.35 |
ENST00000238181.7
|
LGALS8
|
lectin, galactoside-binding, soluble, 8 |
| chr20_+_43211149 | 1.35 |
ENST00000372886.1
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr7_-_138482933 | 1.32 |
ENST00000310018.2
|
ATP6V0A4
|
ATPase, H+ transporting, lysosomal V0 subunit a4 |
| chr11_+_113185292 | 1.30 |
ENST00000429951.1
ENST00000442859.1 ENST00000531164.1 ENST00000529850.1 ENST00000314756.3 ENST00000525965.1 |
TTC12
|
tetratricopeptide repeat domain 12 |
| chr6_+_135502501 | 1.30 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr3_-_47324008 | 1.29 |
ENST00000425853.1
|
KIF9
|
kinesin family member 9 |
| chr14_-_23770683 | 1.29 |
ENST00000561437.1
ENST00000559942.1 ENST00000560913.1 ENST00000559314.1 ENST00000558058.1 |
PPP1R3E
|
protein phosphatase 1, regulatory subunit 3E |
| chr19_+_44645700 | 1.27 |
ENST00000592437.1
|
ZNF234
|
zinc finger protein 234 |
| chr1_+_236687232 | 1.26 |
ENST00000416919.2
ENST00000323938.6 |
LGALS8
|
lectin, galactoside-binding, soluble, 8 |
| chr12_+_56477093 | 1.25 |
ENST00000549672.1
ENST00000415288.2 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr13_+_113812956 | 1.21 |
ENST00000375547.2
ENST00000342783.4 |
PROZ
|
protein Z, vitamin K-dependent plasma glycoprotein |
| chr9_+_133320339 | 1.20 |
ENST00000372394.1
ENST00000372393.3 ENST00000422569.1 |
ASS1
|
argininosuccinate synthase 1 |
| chr2_-_62733476 | 1.20 |
ENST00000335390.5
|
TMEM17
|
transmembrane protein 17 |
| chr6_+_56954867 | 1.19 |
ENST00000370708.4
ENST00000370702.1 |
ZNF451
|
zinc finger protein 451 |
| chr11_+_113185533 | 1.18 |
ENST00000393020.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr19_+_11455900 | 1.16 |
ENST00000588790.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr9_+_2015335 | 1.16 |
ENST00000349721.2
ENST00000357248.2 ENST00000450198.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr14_-_24911971 | 1.16 |
ENST00000555365.1
ENST00000399395.3 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr7_+_33169142 | 1.16 |
ENST00000242067.6
ENST00000350941.3 ENST00000396127.2 ENST00000355070.2 ENST00000354265.4 ENST00000425508.2 |
BBS9
|
Bardet-Biedl syndrome 9 |
| chr1_+_210502238 | 1.15 |
ENST00000545154.1
ENST00000537898.1 ENST00000391905.3 ENST00000545781.1 ENST00000261458.3 ENST00000308852.6 |
HHAT
|
hedgehog acyltransferase |
| chr20_-_43883197 | 1.14 |
ENST00000338380.2
|
SLPI
|
secretory leukocyte peptidase inhibitor |
| chr1_+_202830876 | 1.13 |
ENST00000456105.2
|
RP11-480I12.7
|
RP11-480I12.7 |
| chr19_+_37837185 | 1.13 |
ENST00000541583.2
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr13_+_42031679 | 1.12 |
ENST00000379359.3
|
RGCC
|
regulator of cell cycle |
| chr2_+_217524323 | 1.12 |
ENST00000456764.1
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr16_-_3493528 | 1.10 |
ENST00000301744.4
|
ZNF597
|
zinc finger protein 597 |
| chr1_+_2487800 | 1.08 |
ENST00000355716.4
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr1_-_20126365 | 1.07 |
ENST00000294543.6
ENST00000375122.2 |
TMCO4
|
transmembrane and coiled-coil domains 4 |
| chr14_-_76447494 | 1.07 |
ENST00000238682.3
|
TGFB3
|
transforming growth factor, beta 3 |
| chr16_-_67450325 | 1.05 |
ENST00000348579.2
|
ZDHHC1
|
zinc finger, DHHC-type containing 1 |
| chr19_+_1041212 | 1.05 |
ENST00000433129.1
|
ABCA7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr8_-_143857402 | 1.05 |
ENST00000523332.1
|
LYNX1
|
Ly6/neurotoxin 1 |
| chr2_-_237416071 | 1.05 |
ENST00000309507.5
ENST00000431676.2 |
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr7_-_102715172 | 1.04 |
ENST00000456695.1
ENST00000455112.2 ENST00000440067.1 |
FBXL13
|
F-box and leucine-rich repeat protein 13 |
| chr19_+_1285890 | 1.04 |
ENST00000344663.3
|
MUM1
|
melanoma associated antigen (mutated) 1 |
| chr1_+_160085501 | 1.04 |
ENST00000361216.3
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr20_-_32308028 | 1.03 |
ENST00000409299.3
ENST00000217398.3 ENST00000344022.3 |
PXMP4
|
peroxisomal membrane protein 4, 24kDa |
| chr7_+_99816859 | 1.02 |
ENST00000317271.2
|
PVRIG
|
poliovirus receptor related immunoglobulin domain containing |
| chr2_+_99771418 | 1.02 |
ENST00000393473.2
ENST00000393477.3 ENST00000393474.3 ENST00000340066.1 ENST00000393471.2 ENST00000449211.1 ENST00000434566.1 ENST00000410042.1 |
LIPT1
MRPL30
|
lipoyltransferase 1 39S ribosomal protein L30, mitochondrial |
| chr15_-_43882353 | 1.01 |
ENST00000453080.1
ENST00000360301.4 ENST00000360135.4 ENST00000417085.1 ENST00000431962.1 ENST00000334933.4 ENST00000381879.4 ENST00000420765.1 |
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr11_+_1855645 | 1.00 |
ENST00000381968.3
ENST00000381978.3 |
SYT8
|
synaptotagmin VIII |
| chr1_+_2518202 | 1.00 |
ENST00000419916.2
ENST00000378424.4 ENST00000537325.1 ENST00000378427.1 ENST00000378425.5 ENST00000444521.2 ENST00000465233.1 ENST00000498083.1 |
FAM213B
|
family with sequence similarity 213, member B |
| chr8_-_27468842 | 1.00 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr19_+_49617581 | 1.00 |
ENST00000391864.3
|
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr15_+_66874502 | 0.99 |
ENST00000558797.1
|
RP11-321F6.1
|
HCG2003567; Uncharacterized protein |
| chr12_-_58220078 | 0.99 |
ENST00000549039.1
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr6_-_83775489 | 0.98 |
ENST00000369747.3
|
UBE3D
|
ubiquitin protein ligase E3D |
| chr1_+_15671919 | 0.98 |
ENST00000314668.9
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr16_+_21244986 | 0.98 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr6_+_3000218 | 0.97 |
ENST00000380441.1
ENST00000380455.4 ENST00000380454.4 |
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr5_-_131826457 | 0.96 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr11_+_103907308 | 0.96 |
ENST00000302259.3
|
DDI1
|
DNA-damage inducible 1 homolog 1 (S. cerevisiae) |
| chr19_+_10197463 | 0.95 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr3_-_46000064 | 0.95 |
ENST00000433878.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr16_+_4838412 | 0.94 |
ENST00000589327.1
|
SMIM22
|
small integral membrane protein 22 |
| chr13_+_27131887 | 0.94 |
ENST00000335327.5
|
WASF3
|
WAS protein family, member 3 |
| chr12_-_90049828 | 0.94 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr11_-_3400442 | 0.94 |
ENST00000429541.2
ENST00000532539.1 |
ZNF195
|
zinc finger protein 195 |
| chr22_+_45072958 | 0.93 |
ENST00000403581.1
|
PRR5
|
proline rich 5 (renal) |
| chr12_-_90049878 | 0.93 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr22_+_45072925 | 0.93 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr2_+_12857043 | 0.91 |
ENST00000381465.2
|
TRIB2
|
tribbles pseudokinase 2 |
| chr18_-_712544 | 0.91 |
ENST00000340116.7
ENST00000539164.1 ENST00000580982.1 |
ENOSF1
|
enolase superfamily member 1 |
| chr3_-_47324060 | 0.91 |
ENST00000452770.2
|
KIF9
|
kinesin family member 9 |
| chr4_-_84256024 | 0.90 |
ENST00000311412.5
|
HPSE
|
heparanase |
| chr12_-_56882136 | 0.89 |
ENST00000311966.4
|
GLS2
|
glutaminase 2 (liver, mitochondrial) |
| chr1_+_94883991 | 0.89 |
ENST00000370214.4
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr20_+_19867150 | 0.89 |
ENST00000255006.6
|
RIN2
|
Ras and Rab interactor 2 |
| chrX_-_77395186 | 0.89 |
ENST00000341864.5
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr2_+_14772810 | 0.88 |
ENST00000295092.2
ENST00000331243.4 |
FAM84A
|
family with sequence similarity 84, member A |
| chr19_-_12777509 | 0.86 |
ENST00000221363.4
ENST00000598876.1 ENST00000456935.2 ENST00000486847.2 |
MAN2B1
|
mannosidase, alpha, class 2B, member 1 |
| chr3_+_48507621 | 0.85 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr17_-_38256973 | 0.84 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr17_+_6659153 | 0.84 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr8_+_21911054 | 0.84 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr20_-_40247133 | 0.84 |
ENST00000373233.3
ENST00000309279.7 |
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr14_+_81421710 | 0.83 |
ENST00000342443.6
|
TSHR
|
thyroid stimulating hormone receptor |
| chr9_-_139839064 | 0.83 |
ENST00000325285.3
ENST00000428398.1 |
FBXW5
|
F-box and WD repeat domain containing 5 |
| chr12_-_55367361 | 0.82 |
ENST00000532804.1
ENST00000531122.1 ENST00000533446.1 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr18_+_48556470 | 0.81 |
ENST00000589076.1
ENST00000590061.1 ENST00000591914.1 ENST00000342988.3 |
SMAD4
|
SMAD family member 4 |
| chr14_+_55595762 | 0.81 |
ENST00000254301.9
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr3_-_125820348 | 0.80 |
ENST00000509064.1
ENST00000508835.1 |
SLC41A3
|
solute carrier family 41, member 3 |
| chr8_-_27468945 | 0.80 |
ENST00000405140.3
|
CLU
|
clusterin |
| chr1_+_160370344 | 0.79 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr11_+_66247880 | 0.78 |
ENST00000360510.2
ENST00000453114.1 ENST00000541961.1 ENST00000532019.1 ENST00000526515.1 ENST00000530165.1 ENST00000533725.1 |
DPP3
|
dipeptidyl-peptidase 3 |
| chr8_-_57232656 | 0.78 |
ENST00000396721.2
|
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5 |
| chr14_+_103589789 | 0.76 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr10_+_88854926 | 0.76 |
ENST00000298784.1
ENST00000298786.4 |
FAM35A
|
family with sequence similarity 35, member A |
| chr16_+_29973351 | 0.75 |
ENST00000602948.1
ENST00000279396.6 ENST00000575829.2 ENST00000561899.2 |
TMEM219
|
transmembrane protein 219 |
| chr4_-_681114 | 0.75 |
ENST00000503156.1
|
MFSD7
|
major facilitator superfamily domain containing 7 |
| chr6_-_30080876 | 0.75 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr9_+_124030338 | 0.75 |
ENST00000449773.1
ENST00000432226.1 ENST00000436847.1 ENST00000394353.2 ENST00000449733.1 ENST00000412819.1 ENST00000341272.2 ENST00000373808.2 |
GSN
|
gelsolin |
| chr3_+_169629354 | 0.75 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr1_+_14075865 | 0.74 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr6_+_131894284 | 0.74 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr17_-_41466555 | 0.74 |
ENST00000586231.1
|
LINC00910
|
long intergenic non-protein coding RNA 910 |
| chr1_-_43751230 | 0.74 |
ENST00000523677.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr7_-_99097863 | 0.73 |
ENST00000426306.2
ENST00000337673.6 |
ZNF394
|
zinc finger protein 394 |
| chr9_-_79307096 | 0.73 |
ENST00000376717.2
ENST00000223609.6 ENST00000443509.2 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr11_+_64009072 | 0.73 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr20_+_30458431 | 0.72 |
ENST00000375938.4
ENST00000535842.1 ENST00000310998.4 ENST00000375921.2 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr16_+_84209738 | 0.72 |
ENST00000564928.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr22_+_30163340 | 0.72 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr14_-_53619816 | 0.72 |
ENST00000323669.5
ENST00000395606.1 ENST00000357758.3 |
DDHD1
|
DDHD domain containing 1 |
| chr18_+_74240610 | 0.72 |
ENST00000578092.1
ENST00000578613.1 ENST00000583578.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr5_-_133968529 | 0.72 |
ENST00000402673.2
|
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr10_+_81838411 | 0.71 |
ENST00000372281.3
ENST00000372277.3 ENST00000372275.1 ENST00000372274.1 |
TMEM254
|
transmembrane protein 254 |
| chr4_-_23735183 | 0.71 |
ENST00000507666.1
|
RP11-380P13.2
|
RP11-380P13.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HNF4G
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 19.6 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 1.9 | 9.7 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 1.8 | 9.2 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 1.6 | 4.8 | GO:0033214 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 1.6 | 6.4 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 1.2 | 3.5 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 1.1 | 5.7 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 1.1 | 5.4 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 1.1 | 3.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 1.0 | 2.9 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.9 | 5.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.8 | 3.1 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.7 | 4.8 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.6 | 12.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.6 | 1.7 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.5 | 1.4 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.4 | 1.3 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.4 | 2.5 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.4 | 0.4 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.4 | 1.6 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.4 | 1.2 | GO:0042704 | detection of oxygen(GO:0003032) uterine wall breakdown(GO:0042704) |
| 0.3 | 1.0 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.3 | 2.0 | GO:0043366 | beta selection(GO:0043366) |
| 0.3 | 1.0 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.3 | 0.3 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.3 | 1.6 | GO:2000051 | Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 0.9 | GO:0031643 | positive regulation of myelination(GO:0031643) positive regulation of neurological system process(GO:0031646) |
| 0.3 | 1.5 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.3 | 1.8 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 0.3 | 0.8 | GO:0060086 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.3 | 1.1 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.3 | 1.4 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) malonyl-CoA metabolic process(GO:2001293) |
| 0.3 | 0.8 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.3 | 0.8 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.2 | 1.9 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.2 | 1.0 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.2 | 3.0 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 0.8 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.2 | 2.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 0.6 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.2 | 0.9 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.2 | 0.9 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 1.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.2 | 2.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.2 | 0.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 0.2 | GO:2000506 | negative regulation of energy homeostasis(GO:2000506) |
| 0.2 | 0.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 0.5 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.2 | 0.5 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.1 | 0.7 | GO:0044856 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.4 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.7 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 0.1 | 0.8 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 1.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 1.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.4 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 1.0 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 2.3 | GO:0086027 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.1 | 0.4 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.1 | 1.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.1 | 3.6 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.1 | 0.3 | GO:0032632 | interleukin-3 production(GO:0032632) cadmium ion homeostasis(GO:0055073) divalent metal ion export(GO:0070839) |
| 0.1 | 1.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 2.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.1 | GO:1902908 | regulation of melanosome transport(GO:1902908) |
| 0.1 | 1.6 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 2.6 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 7.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.9 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.9 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.3 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.8 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 1.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.3 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.1 | 0.3 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.1 | 3.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.3 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 0.5 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.3 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.3 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.6 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.1 | 0.3 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.4 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.7 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 1.1 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 0.7 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.6 | GO:0006489 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.1 | 0.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 0.7 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.3 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 1.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 2.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.3 | GO:0000050 | urea cycle(GO:0000050) |
| 0.1 | 1.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.3 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.1 | 0.5 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.6 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 | 0.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.3 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.2 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.1 | 0.2 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.5 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.1 | 1.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.7 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 0.3 | GO:0060611 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.1 | 1.3 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.1 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 0.4 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.3 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.1 | 0.5 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 3.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 4.3 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.1 | 1.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.4 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 1.7 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 3.1 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.1 | 0.3 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.5 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 2.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.5 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) |
| 0.0 | 1.5 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 1.6 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.3 | GO:0016139 | glycoside catabolic process(GO:0016139) glycolipid catabolic process(GO:0019377) |
| 0.0 | 0.6 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 1.5 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.4 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 1.0 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.3 | GO:0044598 | polyketide metabolic process(GO:0030638) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 3.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 1.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.7 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:1904808 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:0072757 | cellular response to camptothecin(GO:0072757) response to camptothecin(GO:1901563) |
| 0.0 | 0.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.9 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.0 | 0.4 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 1.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.2 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.2 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.6 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 2.0 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 2.0 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.2 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.8 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 0.7 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.2 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 2.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.6 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.4 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.7 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 1.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 1.7 | GO:0031016 | pancreas development(GO:0031016) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.4 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.4 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.8 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.4 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 1.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.3 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 1.0 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.3 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.3 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.2 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.2 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.8 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.4 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:2001033 | negative regulation of double-strand break repair via nonhomologous end joining(GO:2001033) |
| 0.0 | 0.3 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 1.3 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.9 | GO:0043525 | positive regulation of neuron apoptotic process(GO:0043525) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 1.0 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.5 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.4 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0071494 | positive regulation of translational initiation in response to stress(GO:0032058) cellular response to UV-C(GO:0071494) |
| 0.0 | 0.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.9 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.4 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.6 | GO:0048806 | genitalia development(GO:0048806) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.5 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.2 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.0 | 0.2 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 0.2 | GO:0030252 | growth hormone secretion(GO:0030252) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.8 | GO:0098536 | deuterosome(GO:0098536) |
| 0.7 | 4.8 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.6 | 0.6 | GO:0097525 | spliceosomal snRNP complex(GO:0097525) |
| 0.5 | 3.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.5 | 2.5 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.4 | 6.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 3.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 0.8 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 0.9 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.2 | 14.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 2.5 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.4 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.8 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 2.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 2.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 1.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 8.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 2.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 1.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 1.6 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.1 | 1.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.0 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 1.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 3.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 6.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 2.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.6 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.4 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 3.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.7 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.7 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 0.3 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.3 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.3 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.9 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 2.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 2.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.4 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 1.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 3.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 2.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 3.1 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 7.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 3.0 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 3.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 1.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 1.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 2.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0045171 | intercellular bridge(GO:0045171) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 32.8 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.9 | 5.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.6 | 6.5 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.5 | 1.6 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.4 | 2.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.4 | 1.9 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.3 | 1.0 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 5.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 1.6 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.3 | 3.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 1.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 2.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 2.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.3 | 3.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.3 | 1.0 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.2 | 1.0 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.2 | 1.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 0.9 | GO:0030305 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.2 | 3.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.5 | GO:0019863 | IgE binding(GO:0019863) |
| 0.2 | 2.6 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 1.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 2.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.6 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.4 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 0.8 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.7 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 2.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 5.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.6 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 2.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.4 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 2.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 4.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.3 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 2.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 2.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.4 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.3 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 1.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 2.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.3 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.7 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.3 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.1 | 1.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.8 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.0 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.1 | 1.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 3.0 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 7.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 6.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.4 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 0.4 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 7.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.2 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.1 | 1.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 1.0 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 3.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 0.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.5 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 2.8 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.1 | 0.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.7 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.4 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.1 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 1.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.9 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.9 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 1.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 1.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 2.3 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.9 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 1.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.9 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.7 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 5.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.9 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 3.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 1.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0008194 | UDP-glycosyltransferase activity(GO:0008194) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.4 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 2.8 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.7 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 1.5 | GO:0016597 | amino acid binding(GO:0016597) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.7 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 5.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 8.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 4.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 2.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 25.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 3.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 2.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 8.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 3.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 3.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 3.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 3.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 3.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 6.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 2.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.8 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.9 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.1 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.2 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |