Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for HOXA10_HOXB9
Z-value: 0.50
Transcription factors associated with HOXA10_HOXB9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
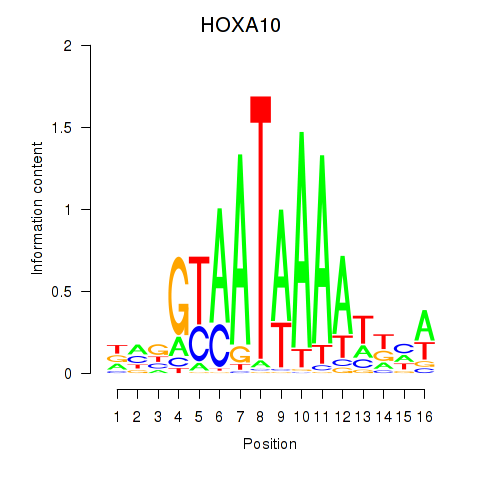
HOXA10
|
ENSG00000253293.3 | homeobox A10 |
|
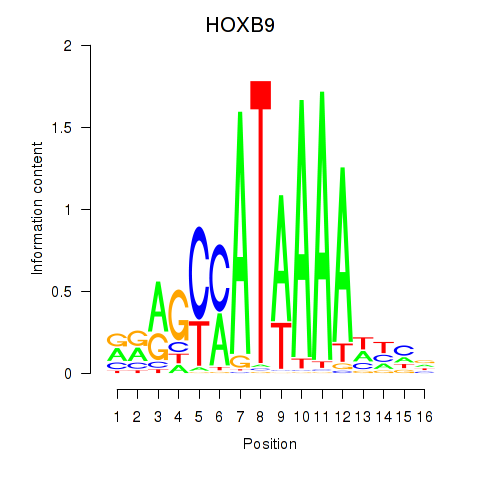
HOXB9
|
ENSG00000170689.8 | homeobox B9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA10 | hg19_v2_chr7_-_27213893_27213954 | -0.61 | 3.6e-04 | Click! |
| HOXB9 | hg19_v2_chr17_-_46703826_46703845 | -0.34 | 6.9e-02 | Click! |
Activity profile of HOXA10_HOXB9 motif
Sorted Z-values of HOXA10_HOXB9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_160231451 | 1.09 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr5_-_88119580 | 0.57 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr20_-_7921090 | 0.54 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr3_+_169629354 | 0.52 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr7_+_35756186 | 0.51 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr12_+_20963632 | 0.48 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr12_-_95945246 | 0.47 |
ENST00000258499.3
|
USP44
|
ubiquitin specific peptidase 44 |
| chr12_+_20963647 | 0.47 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr18_-_19994830 | 0.45 |
ENST00000525417.1
|
CTAGE1
|
cutaneous T-cell lymphoma-associated antigen 1 |
| chr15_+_40886199 | 0.44 |
ENST00000346991.5
ENST00000528975.1 ENST00000527044.1 |
CASC5
|
cancer susceptibility candidate 5 |
| chr14_+_73706308 | 0.41 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr7_-_41742697 | 0.41 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chrX_-_73072534 | 0.40 |
ENST00000429829.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr12_+_64173583 | 0.39 |
ENST00000261234.6
|
TMEM5
|
transmembrane protein 5 |
| chr19_+_21106081 | 0.38 |
ENST00000300540.3
ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chr5_+_98109322 | 0.38 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr15_-_98417780 | 0.36 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr12_-_90049878 | 0.34 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr1_+_244515930 | 0.34 |
ENST00000366537.1
ENST00000308105.4 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr10_+_102106829 | 0.33 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr20_+_4702548 | 0.33 |
ENST00000305817.2
|
PRND
|
prion protein 2 (dublet) |
| chr14_-_25479811 | 0.32 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr12_-_90049828 | 0.32 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr9_-_95640218 | 0.30 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr15_-_58571445 | 0.29 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr22_+_41956767 | 0.29 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr1_+_116654376 | 0.29 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr15_+_67418047 | 0.26 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr9_+_125132803 | 0.26 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr15_+_40886439 | 0.26 |
ENST00000532056.1
ENST00000399668.2 |
CASC5
|
cancer susceptibility candidate 5 |
| chr1_+_46152886 | 0.25 |
ENST00000372025.4
|
TMEM69
|
transmembrane protein 69 |
| chr17_+_48823975 | 0.25 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr4_-_110723194 | 0.25 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr15_-_55541227 | 0.25 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr5_+_68860949 | 0.25 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr11_-_102576537 | 0.24 |
ENST00000260229.4
|
MMP27
|
matrix metallopeptidase 27 |
| chr17_+_4487294 | 0.24 |
ENST00000338859.4
|
SMTNL2
|
smoothelin-like 2 |
| chr12_-_57146095 | 0.24 |
ENST00000550770.1
ENST00000338193.6 |
PRIM1
|
primase, DNA, polypeptide 1 (49kDa) |
| chr6_+_46761118 | 0.24 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr11_+_63606373 | 0.23 |
ENST00000402010.2
ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr4_+_70861647 | 0.23 |
ENST00000246895.4
ENST00000381060.2 |
STATH
|
statherin |
| chr9_+_42671887 | 0.23 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr13_+_108922228 | 0.23 |
ENST00000542136.1
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr15_+_66797627 | 0.22 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr10_+_85933494 | 0.22 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr2_-_216878305 | 0.22 |
ENST00000263268.6
|
MREG
|
melanoregulin |
| chr14_+_79745746 | 0.22 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr1_+_2985726 | 0.21 |
ENST00000511072.1
ENST00000378398.3 ENST00000441472.2 ENST00000442529.2 |
PRDM16
|
PR domain containing 16 |
| chr10_-_73848531 | 0.21 |
ENST00000373109.2
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr10_-_73848086 | 0.21 |
ENST00000536168.1
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr9_-_179018 | 0.21 |
ENST00000431099.2
ENST00000382447.4 ENST00000382389.1 ENST00000377447.3 ENST00000314367.10 ENST00000356521.4 ENST00000382393.1 ENST00000377400.4 |
CBWD1
|
COBW domain containing 1 |
| chr18_-_52989525 | 0.20 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr4_+_71263599 | 0.20 |
ENST00000399575.2
|
PROL1
|
proline rich, lacrimal 1 |
| chr9_+_70856397 | 0.20 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr2_+_161993465 | 0.20 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr6_+_12958137 | 0.20 |
ENST00000457702.2
ENST00000379345.2 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr14_+_56584414 | 0.19 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr15_+_66797455 | 0.19 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr10_-_129691195 | 0.19 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
| chr12_+_70219052 | 0.18 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr1_-_68962805 | 0.18 |
ENST00000370966.5
|
DEPDC1
|
DEP domain containing 1 |
| chr6_+_28249299 | 0.18 |
ENST00000405948.2
|
PGBD1
|
piggyBac transposable element derived 1 |
| chrX_-_80457385 | 0.18 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr6_+_28249332 | 0.17 |
ENST00000259883.3
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr6_+_111408698 | 0.17 |
ENST00000368851.5
|
SLC16A10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr8_-_25281747 | 0.17 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr1_+_153003671 | 0.16 |
ENST00000307098.4
|
SPRR1B
|
small proline-rich protein 1B |
| chr20_+_10015678 | 0.16 |
ENST00000378392.1
ENST00000378380.3 |
ANKEF1
|
ankyrin repeat and EF-hand domain containing 1 |
| chr5_+_34915444 | 0.16 |
ENST00000336767.5
|
BRIX1
|
BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) |
| chr2_+_31456874 | 0.15 |
ENST00000541626.1
|
EHD3
|
EH-domain containing 3 |
| chr5_-_36001108 | 0.15 |
ENST00000333811.4
|
UGT3A1
|
UDP glycosyltransferase 3 family, polypeptide A1 |
| chr1_+_154401791 | 0.15 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr11_-_104916034 | 0.15 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr8_-_81083731 | 0.15 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chr10_+_115614370 | 0.15 |
ENST00000369301.3
|
NHLRC2
|
NHL repeat containing 2 |
| chr18_-_52989217 | 0.15 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr8_+_77593474 | 0.15 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr1_+_110993795 | 0.14 |
ENST00000271331.3
|
PROK1
|
prokineticin 1 |
| chr1_-_161207953 | 0.14 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_-_161208013 | 0.14 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_+_12524965 | 0.14 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr12_-_18243119 | 0.14 |
ENST00000538724.1
ENST00000229002.2 |
RERGL
|
RERG/RAS-like |
| chr5_+_140261703 | 0.13 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr14_+_53173890 | 0.13 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr9_+_134065506 | 0.13 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr8_+_77593448 | 0.13 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr18_+_3252206 | 0.13 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr9_-_69262509 | 0.13 |
ENST00000377449.1
ENST00000382399.4 ENST00000377439.1 ENST00000377441.1 ENST00000377457.5 |
CBWD6
|
COBW domain containing 6 |
| chr21_-_31869451 | 0.13 |
ENST00000334058.2
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr19_+_48497962 | 0.13 |
ENST00000596043.1
ENST00000597519.1 |
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr2_+_102928009 | 0.13 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr19_+_48497901 | 0.13 |
ENST00000339841.2
|
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr3_+_149191723 | 0.12 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr1_+_229440129 | 0.12 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr12_-_43833515 | 0.12 |
ENST00000549670.1
ENST00000395541.2 |
ADAMTS20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr8_-_143859197 | 0.12 |
ENST00000395192.2
|
LYNX1
|
Ly6/neurotoxin 1 |
| chrX_+_41548259 | 0.12 |
ENST00000378138.5
|
GPR34
|
G protein-coupled receptor 34 |
| chr16_-_15180257 | 0.12 |
ENST00000540462.1
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr14_+_53173910 | 0.12 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr12_-_71148357 | 0.12 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr11_+_120973375 | 0.12 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr14_+_79745682 | 0.12 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr2_+_185463093 | 0.12 |
ENST00000302277.6
|
ZNF804A
|
zinc finger protein 804A |
| chr2_+_113885138 | 0.12 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr8_-_81083890 | 0.11 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chr3_+_138340049 | 0.11 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr12_-_71148413 | 0.11 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr20_-_25320367 | 0.11 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chr3_+_133118839 | 0.11 |
ENST00000302334.2
|
BFSP2
|
beaded filament structural protein 2, phakinin |
| chr16_-_80603558 | 0.11 |
ENST00000567317.1
|
RP11-18F14.1
|
RP11-18F14.1 |
| chr18_+_3252265 | 0.11 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr6_+_80816342 | 0.11 |
ENST00000369760.4
ENST00000356489.5 ENST00000320393.6 |
BCKDHB
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr3_+_107602030 | 0.11 |
ENST00000494231.1
|
LINC00636
|
long intergenic non-protein coding RNA 636 |
| chr1_+_84630645 | 0.11 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr2_+_27440229 | 0.11 |
ENST00000264705.4
ENST00000403525.1 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr1_-_161207986 | 0.11 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr4_-_57522598 | 0.11 |
ENST00000553379.2
|
HOPX
|
HOP homeobox |
| chr4_+_71226468 | 0.11 |
ENST00000226460.4
|
SMR3A
|
submaxillary gland androgen regulated protein 3A |
| chr11_-_3400442 | 0.11 |
ENST00000429541.2
ENST00000532539.1 |
ZNF195
|
zinc finger protein 195 |
| chr2_+_161993412 | 0.11 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr8_-_21669826 | 0.11 |
ENST00000517328.1
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr5_+_140514782 | 0.10 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr4_-_57522673 | 0.10 |
ENST00000381255.3
ENST00000317745.7 ENST00000555760.2 ENST00000556614.2 |
HOPX
|
HOP homeobox |
| chr12_+_80603233 | 0.10 |
ENST00000547103.1
ENST00000458043.2 |
OTOGL
|
otogelin-like |
| chr3_+_15045419 | 0.10 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr11_-_3400330 | 0.10 |
ENST00000427810.2
ENST00000005082.9 ENST00000534569.1 ENST00000438262.2 ENST00000528796.1 ENST00000528410.1 ENST00000529678.1 ENST00000354599.6 ENST00000526601.1 ENST00000525502.1 ENST00000533036.1 ENST00000399602.4 |
ZNF195
|
zinc finger protein 195 |
| chr10_-_115614127 | 0.10 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chrX_-_135962876 | 0.10 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr9_+_124413873 | 0.10 |
ENST00000408936.3
|
DAB2IP
|
DAB2 interacting protein |
| chr3_+_138340067 | 0.10 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr9_+_44868935 | 0.09 |
ENST00000448436.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr12_-_18243075 | 0.09 |
ENST00000536890.1
|
RERGL
|
RERG/RAS-like |
| chr5_+_141346385 | 0.09 |
ENST00000513019.1
ENST00000356143.1 |
RNF14
|
ring finger protein 14 |
| chr5_-_125930929 | 0.09 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr9_-_85678043 | 0.09 |
ENST00000376447.3
ENST00000340717.4 |
RASEF
|
RAS and EF-hand domain containing |
| chr18_-_53069419 | 0.09 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr22_-_22292934 | 0.09 |
ENST00000538191.1
ENST00000424647.1 ENST00000407142.1 |
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr5_+_140165876 | 0.09 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr3_+_189507523 | 0.09 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr5_-_139937895 | 0.09 |
ENST00000336283.6
|
SRA1
|
steroid receptor RNA activator 1 |
| chr6_+_158733692 | 0.09 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr15_+_63414760 | 0.09 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr7_-_77427676 | 0.09 |
ENST00000257663.3
|
TMEM60
|
transmembrane protein 60 |
| chr2_-_176046391 | 0.09 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr4_+_146539415 | 0.09 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr8_+_24241969 | 0.09 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr2_+_101591314 | 0.09 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr8_+_132952112 | 0.09 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr3_-_108672609 | 0.08 |
ENST00000393963.3
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr1_-_161207875 | 0.08 |
ENST00000512372.1
ENST00000437437.2 ENST00000442691.2 ENST00000412844.2 ENST00000428574.2 ENST00000505005.1 ENST00000508740.1 ENST00000508387.1 ENST00000504010.1 ENST00000511676.1 ENST00000502985.1 ENST00000367981.3 ENST00000515621.1 ENST00000511944.1 ENST00000511748.1 ENST00000367984.4 ENST00000367985.3 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr3_-_108672742 | 0.08 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr3_+_189507460 | 0.08 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr1_-_86848760 | 0.08 |
ENST00000460698.2
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chrX_-_117119243 | 0.08 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr3_-_100712352 | 0.08 |
ENST00000471714.1
ENST00000284322.5 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chrX_+_73164149 | 0.08 |
ENST00000602938.1
ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chrX_-_100604184 | 0.08 |
ENST00000372902.3
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr12_-_71551868 | 0.08 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr1_+_109102652 | 0.08 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr11_-_114430570 | 0.08 |
ENST00000251921.2
|
NXPE1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr5_-_176433693 | 0.08 |
ENST00000507513.1
ENST00000511320.1 |
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr13_-_47012325 | 0.08 |
ENST00000409879.2
|
KIAA0226L
|
KIAA0226-like |
| chr6_-_109804412 | 0.08 |
ENST00000230122.3
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chr7_-_80141328 | 0.08 |
ENST00000398291.3
|
GNAT3
|
guanine nucleotide binding protein, alpha transducing 3 |
| chr11_-_104972158 | 0.08 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr15_+_78830023 | 0.08 |
ENST00000599596.1
|
AC027228.1
|
HCG2005369; Uncharacterized protein |
| chr4_-_57547870 | 0.08 |
ENST00000381260.3
ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX
|
HOP homeobox |
| chr12_+_57810198 | 0.08 |
ENST00000598001.1
|
AC126614.1
|
HCG1818482; Uncharacterized protein |
| chr11_+_22688150 | 0.07 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr4_-_69536346 | 0.07 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr5_-_16738451 | 0.07 |
ENST00000274203.9
ENST00000515803.1 |
MYO10
|
myosin X |
| chr2_-_152382500 | 0.07 |
ENST00000434685.1
|
NEB
|
nebulin |
| chr6_+_25754927 | 0.07 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr2_-_71062938 | 0.07 |
ENST00000410009.3
|
CD207
|
CD207 molecule, langerin |
| chr12_-_71551652 | 0.07 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chrX_+_55744166 | 0.07 |
ENST00000374941.4
ENST00000414239.1 |
RRAGB
|
Ras-related GTP binding B |
| chr10_-_103599591 | 0.07 |
ENST00000348850.5
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr9_-_104198042 | 0.07 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr3_+_189507432 | 0.07 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr5_+_43121698 | 0.07 |
ENST00000505606.2
ENST00000509634.1 ENST00000509341.1 |
ZNF131
|
zinc finger protein 131 |
| chr14_-_68000442 | 0.07 |
ENST00000554278.1
|
TMEM229B
|
transmembrane protein 229B |
| chr6_-_52859046 | 0.06 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr20_-_34287103 | 0.06 |
ENST00000374085.1
ENST00000419569.1 |
NFS1
|
NFS1 cysteine desulfurase |
| chr6_-_8102714 | 0.06 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chrX_-_122756660 | 0.06 |
ENST00000441692.1
|
THOC2
|
THO complex 2 |
| chr13_+_24844819 | 0.06 |
ENST00000399949.2
|
SPATA13
|
spermatogenesis associated 13 |
| chrX_+_55744228 | 0.06 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr3_+_89156799 | 0.06 |
ENST00000452448.2
ENST00000494014.1 |
EPHA3
|
EPH receptor A3 |
| chr3_-_38992052 | 0.06 |
ENST00000302328.3
ENST00000450244.1 ENST00000444237.2 |
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit |
| chr2_+_109204909 | 0.06 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr4_+_37455536 | 0.06 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr6_-_83903600 | 0.06 |
ENST00000506587.1
ENST00000507554.1 |
PGM3
|
phosphoglucomutase 3 |
| chr2_+_32390925 | 0.06 |
ENST00000440718.1
ENST00000379343.2 ENST00000282587.5 ENST00000435660.1 ENST00000538303.1 ENST00000357055.3 ENST00000406369.1 |
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr19_+_21265028 | 0.06 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714 |
| chr1_-_220263096 | 0.06 |
ENST00000463953.1
ENST00000354807.3 ENST00000414869.2 ENST00000498237.2 ENST00000498791.2 ENST00000544404.1 ENST00000480959.2 ENST00000322067.7 |
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr6_+_131958436 | 0.06 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr9_-_114521783 | 0.06 |
ENST00000394779.3
ENST00000394777.4 |
C9orf84
|
chromosome 9 open reading frame 84 |
| chr16_+_71560154 | 0.06 |
ENST00000539698.3
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr9_+_34652164 | 0.06 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr13_+_24144509 | 0.06 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA10_HOXB9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.4 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.3 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.7 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.5 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.3 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0031664 | positive regulation of response to biotic stimulus(GO:0002833) lipopolysaccharide-mediated signaling pathway(GO:0031663) regulation of lipopolysaccharide-mediated signaling pathway(GO:0031664) positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.4 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.0 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.0 | 0.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.3 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.0 | GO:0019348 | dolichol metabolic process(GO:0019348) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.3 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.2 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.5 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.3 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.2 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |