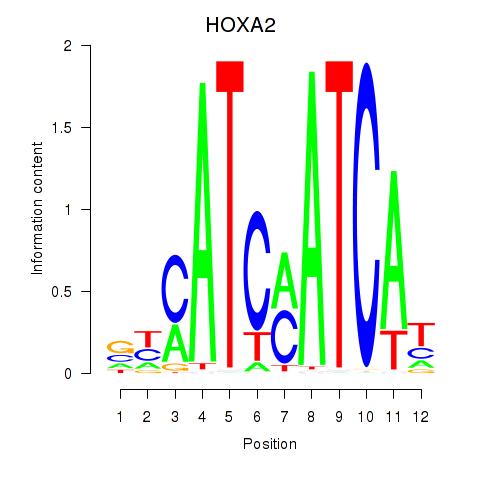
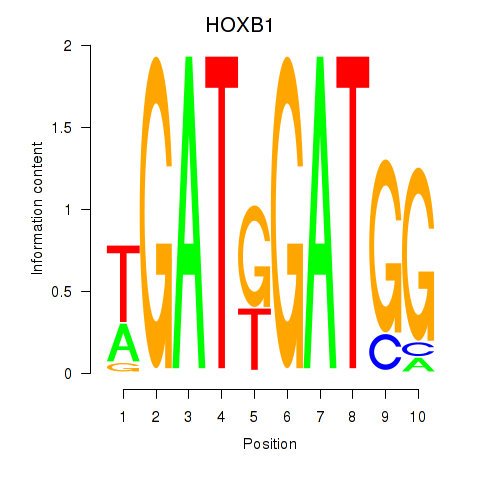
Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for HOXA2_HOXB1
Z-value: 0.59
Transcription factors associated with HOXA2_HOXB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA2
|
ENSG00000105996.5 | homeobox A2 |
|
HOXB1
|
ENSG00000120094.6 | homeobox B1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA2 | hg19_v2_chr7_-_27142290_27142430 | 0.10 | 6.2e-01 | Click! |
| HOXB1 | hg19_v2_chr17_-_46608272_46608385 | -0.05 | 7.8e-01 | Click! |
Activity profile of HOXA2_HOXB1 motif
Sorted Z-values of HOXA2_HOXB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_151561085 | 2.71 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr6_+_151561506 | 2.49 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr11_-_2162162 | 1.36 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr2_-_190044480 | 1.26 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr11_-_111782484 | 1.12 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr15_-_74501360 | 1.05 |
ENST00000323940.5
|
STRA6
|
stimulated by retinoic acid 6 |
| chr1_-_205391178 | 1.05 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr4_-_90757364 | 0.88 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_-_111782696 | 0.87 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr3_-_51813009 | 0.85 |
ENST00000398780.3
|
IQCF6
|
IQ motif containing F6 |
| chr4_-_90756769 | 0.80 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr17_+_37824700 | 0.77 |
ENST00000581428.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr17_-_46623441 | 0.74 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr10_-_79397202 | 0.73 |
ENST00000372437.1
ENST00000372408.2 ENST00000372403.4 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr10_-_79397316 | 0.72 |
ENST00000372421.5
ENST00000457953.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr10_-_100995540 | 0.70 |
ENST00000370546.1
ENST00000404542.1 |
HPSE2
|
heparanase 2 |
| chr1_-_182641367 | 0.66 |
ENST00000508450.1
|
RGS8
|
regulator of G-protein signaling 8 |
| chr12_+_21168630 | 0.66 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chrX_-_30993201 | 0.62 |
ENST00000288422.2
ENST00000378932.2 |
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr10_-_79397479 | 0.59 |
ENST00000404771.3
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr17_+_37824217 | 0.56 |
ENST00000394246.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr12_+_7023491 | 0.56 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr15_-_64673630 | 0.55 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr17_+_37824411 | 0.54 |
ENST00000269582.2
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr19_-_51512804 | 0.52 |
ENST00000594211.1
ENST00000376832.4 |
KLK9
|
kallikrein-related peptidase 9 |
| chr8_-_143961236 | 0.50 |
ENST00000377675.3
ENST00000517471.1 ENST00000292427.4 |
CYP11B1
|
cytochrome P450, family 11, subfamily B, polypeptide 1 |
| chr5_+_167181917 | 0.49 |
ENST00000519204.1
|
TENM2
|
teneurin transmembrane protein 2 |
| chr17_+_7758374 | 0.43 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr1_-_43833628 | 0.42 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr17_+_39975544 | 0.42 |
ENST00000544340.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr15_-_95870310 | 0.42 |
ENST00000508732.2
|
CTD-2536I1.1
|
CTD-2536I1.1 |
| chr7_-_143956815 | 0.40 |
ENST00000493325.1
|
OR2A7
|
olfactory receptor, family 2, subfamily A, member 7 |
| chr17_+_39975455 | 0.40 |
ENST00000455106.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr1_-_182642017 | 0.40 |
ENST00000367557.4
ENST00000258302.4 |
RGS8
|
regulator of G-protein signaling 8 |
| chr10_-_100995603 | 0.39 |
ENST00000370552.3
ENST00000370549.1 |
HPSE2
|
heparanase 2 |
| chr11_+_57365150 | 0.38 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr7_-_144107320 | 0.37 |
ENST00000483238.1
ENST00000467773.1 |
NOBOX
|
NOBOX oogenesis homeobox |
| chr2_-_154335300 | 0.36 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr12_+_101988627 | 0.36 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr9_+_132099158 | 0.35 |
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr10_-_98945515 | 0.34 |
ENST00000371070.4
|
SLIT1
|
slit homolog 1 (Drosophila) |
| chr17_-_27418537 | 0.34 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr12_-_15038779 | 0.33 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr9_-_127269661 | 0.32 |
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr20_+_52105495 | 0.32 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr12_+_57610562 | 0.31 |
ENST00000349394.5
|
NXPH4
|
neurexophilin 4 |
| chr14_+_68189190 | 0.29 |
ENST00000539142.1
|
RDH12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis) |
| chr11_-_126870655 | 0.29 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr16_+_30080857 | 0.28 |
ENST00000565355.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr12_+_101988774 | 0.28 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr7_-_86595190 | 0.28 |
ENST00000398276.2
ENST00000416314.1 ENST00000425689.1 |
KIAA1324L
|
KIAA1324-like |
| chrX_+_99899180 | 0.26 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr17_+_36584662 | 0.25 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr1_+_178062855 | 0.25 |
ENST00000448150.3
|
RASAL2
|
RAS protein activator like 2 |
| chr22_-_39151463 | 0.25 |
ENST00000405510.1
ENST00000433561.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr2_-_197226875 | 0.24 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr9_+_124329336 | 0.24 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr3_+_189507432 | 0.24 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr3_+_189507523 | 0.23 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr7_-_150864635 | 0.23 |
ENST00000297537.4
|
GBX1
|
gastrulation brain homeobox 1 |
| chr12_-_28124903 | 0.23 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr10_-_105615164 | 0.22 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr1_+_243419306 | 0.22 |
ENST00000355875.4
ENST00000391846.1 ENST00000366541.3 ENST00000343783.6 |
SDCCAG8
|
serologically defined colon cancer antigen 8 |
| chr18_+_56887381 | 0.22 |
ENST00000256857.2
ENST00000529320.2 ENST00000420468.2 |
GRP
|
gastrin-releasing peptide |
| chr7_-_27170352 | 0.21 |
ENST00000428284.2
ENST00000360046.5 |
HOXA4
|
homeobox A4 |
| chr17_+_43239191 | 0.21 |
ENST00000589230.1
|
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr14_-_81893734 | 0.21 |
ENST00000555447.1
|
STON2
|
stonin 2 |
| chr17_-_37353950 | 0.21 |
ENST00000394310.3
ENST00000394303.3 ENST00000344140.5 |
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr15_-_64673665 | 0.20 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr5_-_179045199 | 0.20 |
ENST00000523921.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr8_+_143530791 | 0.19 |
ENST00000517894.1
|
BAI1
|
brain-specific angiogenesis inhibitor 1 |
| chr12_+_56414851 | 0.18 |
ENST00000547167.1
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr1_+_68150744 | 0.15 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr8_-_67874805 | 0.15 |
ENST00000563496.1
|
TCF24
|
transcription factor 24 |
| chr17_-_1588101 | 0.14 |
ENST00000577001.1
ENST00000572621.1 ENST00000304992.6 |
PRPF8
|
pre-mRNA processing factor 8 |
| chr7_+_20370746 | 0.14 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr5_+_162864575 | 0.13 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr12_-_28125638 | 0.13 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr5_+_145826867 | 0.13 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr17_-_57229155 | 0.13 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr8_+_23386557 | 0.13 |
ENST00000523930.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr19_+_35849723 | 0.13 |
ENST00000594310.1
|
FFAR3
|
free fatty acid receptor 3 |
| chr16_+_25703274 | 0.12 |
ENST00000331351.5
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr2_+_242089833 | 0.12 |
ENST00000404405.3
ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7
|
protein phosphatase 1, regulatory subunit 7 |
| chr7_-_14029283 | 0.11 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr17_+_79495397 | 0.11 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr10_+_24755416 | 0.11 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr9_-_73029540 | 0.11 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr4_-_102267953 | 0.11 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr3_+_150264555 | 0.10 |
ENST00000406576.3
ENST00000482093.1 ENST00000273435.5 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr15_-_74501310 | 0.10 |
ENST00000423167.2
ENST00000432245.2 |
STRA6
|
stimulated by retinoic acid 6 |
| chr9_-_36276966 | 0.10 |
ENST00000543356.2
ENST00000396594.3 |
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr3_+_148545586 | 0.10 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr20_+_42574317 | 0.10 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr3_+_150264458 | 0.09 |
ENST00000487799.1
ENST00000460851.1 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr19_+_13051206 | 0.09 |
ENST00000586760.1
|
CALR
|
calreticulin |
| chr10_-_98945677 | 0.09 |
ENST00000266058.4
ENST00000371041.3 |
SLIT1
|
slit homolog 1 (Drosophila) |
| chr6_+_33387830 | 0.09 |
ENST00000293748.5
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1 |
| chr6_+_33388013 | 0.09 |
ENST00000449372.2
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1 |
| chr10_+_18689637 | 0.09 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr7_-_14029515 | 0.08 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr17_+_48609903 | 0.08 |
ENST00000268933.3
|
EPN3
|
epsin 3 |
| chr12_+_57482877 | 0.08 |
ENST00000342556.6
ENST00000357680.4 |
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr1_-_109584768 | 0.07 |
ENST00000357672.3
|
WDR47
|
WD repeat domain 47 |
| chr7_-_100881109 | 0.07 |
ENST00000308344.5
|
CLDN15
|
claudin 15 |
| chr7_+_26591441 | 0.07 |
ENST00000420912.1
ENST00000457000.1 ENST00000430426.1 |
AC004947.2
|
AC004947.2 |
| chr8_+_23386305 | 0.07 |
ENST00000519973.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr7_-_27142290 | 0.07 |
ENST00000222718.5
|
HOXA2
|
homeobox A2 |
| chr12_-_86650077 | 0.07 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr20_+_34802295 | 0.07 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr2_+_68961934 | 0.07 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr12_+_10658489 | 0.07 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr6_+_50061315 | 0.06 |
ENST00000415106.1
|
RP11-397G17.1
|
RP11-397G17.1 |
| chr1_-_108507631 | 0.06 |
ENST00000527011.1
ENST00000370056.4 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr1_-_109584716 | 0.06 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chr2_-_145277882 | 0.06 |
ENST00000465070.1
ENST00000444559.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr1_-_109584608 | 0.06 |
ENST00000400794.3
ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47
|
WD repeat domain 47 |
| chr12_-_57472522 | 0.05 |
ENST00000379391.3
ENST00000300128.4 |
TMEM194A
|
transmembrane protein 194A |
| chr9_-_101471479 | 0.05 |
ENST00000259455.2
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr2_+_68961905 | 0.05 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr6_-_112575912 | 0.04 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chrX_-_110655391 | 0.04 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chrX_+_41583408 | 0.04 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr20_+_34742650 | 0.04 |
ENST00000373945.1
ENST00000338074.2 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr7_-_27135591 | 0.04 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr14_+_22985251 | 0.03 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27 |
| chr19_+_52074502 | 0.03 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr20_+_30467600 | 0.02 |
ENST00000375934.4
ENST00000375922.4 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr9_+_113431059 | 0.02 |
ENST00000416899.2
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chr12_-_88423164 | 0.02 |
ENST00000298699.2
ENST00000550553.1 |
C12orf50
|
chromosome 12 open reading frame 50 |
| chr9_-_26947453 | 0.02 |
ENST00000397292.3
|
PLAA
|
phospholipase A2-activating protein |
| chr15_+_48009541 | 0.01 |
ENST00000536845.2
ENST00000558816.1 |
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr9_-_73477826 | 0.01 |
ENST00000396285.1
ENST00000396292.4 ENST00000396280.5 ENST00000358082.3 ENST00000408909.2 ENST00000377097.3 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr3_+_191046810 | 0.00 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr6_+_29426230 | 0.00 |
ENST00000442615.1
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr12_-_58135903 | 0.00 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr2_-_49381572 | 0.00 |
ENST00000454032.1
ENST00000304421.4 |
FSHR
|
follicle stimulating hormone receptor |
| chrX_-_45629661 | 0.00 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr16_-_49890016 | 0.00 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chrX_-_47509887 | 0.00 |
ENST00000247161.3
ENST00000592066.1 ENST00000376983.3 |
ELK1
|
ELK1, member of ETS oncogene family |
| chr14_-_21516590 | 0.00 |
ENST00000555026.1
|
NDRG2
|
NDRG family member 2 |
| chrY_+_26997726 | 0.00 |
ENST00000382296.2
|
DAZ4
|
deleted in azoospermia 4 |
| chr11_+_68228186 | 0.00 |
ENST00000393799.2
ENST00000393800.2 ENST00000528635.1 ENST00000533127.1 ENST00000529907.1 ENST00000529344.1 ENST00000534534.1 ENST00000524845.1 ENST00000265637.4 ENST00000524904.1 ENST00000393801.3 ENST00000265636.5 ENST00000529710.1 |
PPP6R3
|
protein phosphatase 6, regulatory subunit 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA2_HOXB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.4 | 1.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 5.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.3 | 1.7 | GO:0051620 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.3 | 0.8 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.3 | 0.8 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.2 | 1.2 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.2 | 2.0 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.2 | 1.4 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 2.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.4 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.4 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.1 | 0.3 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.5 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.2 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 1.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.2 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.3 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.0 | 0.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.8 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 1.1 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 2.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 2.0 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 2.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 5.1 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.3 | 1.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.3 | 5.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.3 | 1.1 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.3 | 2.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 0.5 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 2.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 1.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 1.2 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 1.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |