Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
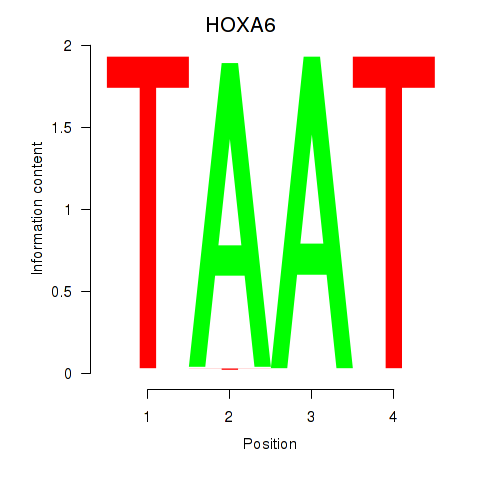
Results for HOXA6
Z-value: 0.42
Transcription factors associated with HOXA6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA6
|
ENSG00000106006.6 | homeobox A6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA6 | hg19_v2_chr7_-_27187393_27187393 | -0.09 | 6.3e-01 | Click! |
Activity profile of HOXA6 motif
Sorted Z-values of HOXA6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_27183263 | 0.94 |
ENST00000222726.3
|
HOXA5
|
homeobox A5 |
| chr4_-_41884620 | 0.63 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr2_+_162272605 | 0.55 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr17_-_38859996 | 0.54 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr1_-_152131703 | 0.39 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr8_-_133123406 | 0.33 |
ENST00000434736.2
|
HHLA1
|
HERV-H LTR-associating 1 |
| chr13_-_95131923 | 0.30 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr12_-_89746173 | 0.29 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr12_-_13248598 | 0.28 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr2_-_29297127 | 0.27 |
ENST00000331664.5
|
C2orf71
|
chromosome 2 open reading frame 71 |
| chrX_+_135279179 | 0.26 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr8_+_104831472 | 0.25 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr7_-_27142290 | 0.25 |
ENST00000222718.5
|
HOXA2
|
homeobox A2 |
| chr14_-_54423529 | 0.25 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr18_+_28956740 | 0.24 |
ENST00000308128.4
ENST00000359747.4 |
DSG4
|
desmoglein 4 |
| chr15_+_71839566 | 0.23 |
ENST00000357769.4
|
THSD4
|
thrombospondin, type I, domain containing 4 |
| chr20_-_50418972 | 0.23 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr3_+_141105705 | 0.23 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr20_-_50419055 | 0.23 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr20_-_50418947 | 0.23 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr7_-_27205136 | 0.22 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chr15_+_93443419 | 0.21 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr7_+_107110488 | 0.21 |
ENST00000304402.4
|
GPR22
|
G protein-coupled receptor 22 |
| chr6_-_9933500 | 0.21 |
ENST00000492169.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr10_+_115312766 | 0.20 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr14_-_57272366 | 0.20 |
ENST00000554788.1
ENST00000554845.1 ENST00000408990.3 |
OTX2
|
orthodenticle homeobox 2 |
| chr14_-_92413727 | 0.19 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr9_+_2717502 | 0.19 |
ENST00000382082.3
|
KCNV2
|
potassium channel, subfamily V, member 2 |
| chr5_-_1882858 | 0.18 |
ENST00000511126.1
ENST00000231357.2 |
IRX4
|
iroquois homeobox 4 |
| chr17_-_38911580 | 0.18 |
ENST00000312150.4
|
KRT25
|
keratin 25 |
| chr2_-_177502659 | 0.18 |
ENST00000295549.4
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr3_-_114477787 | 0.17 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_+_171034646 | 0.17 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chrX_+_16668278 | 0.17 |
ENST00000380200.3
|
S100G
|
S100 calcium binding protein G |
| chr10_+_24528108 | 0.17 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chrX_+_135251835 | 0.17 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr18_-_24237339 | 0.17 |
ENST00000580191.1
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr6_+_108487245 | 0.16 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr4_-_116034979 | 0.16 |
ENST00000264363.2
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr5_-_160973649 | 0.16 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr17_-_56082455 | 0.15 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr3_-_54962100 | 0.15 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr6_-_108145499 | 0.14 |
ENST00000369020.3
ENST00000369022.2 |
SCML4
|
sex comb on midleg-like 4 (Drosophila) |
| chr3_-_114477962 | 0.14 |
ENST00000471418.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr9_+_135457530 | 0.14 |
ENST00000263610.2
|
BARHL1
|
BarH-like homeobox 1 |
| chr9_-_73483958 | 0.14 |
ENST00000377101.1
ENST00000377106.1 ENST00000360823.2 ENST00000377105.1 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chrX_+_135251783 | 0.14 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr20_+_59654146 | 0.14 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr5_-_61031495 | 0.13 |
ENST00000506550.1
ENST00000512882.2 |
CTD-2170G1.2
|
CTD-2170G1.2 |
| chr16_+_89334512 | 0.13 |
ENST00000602042.1
|
AC137932.1
|
AC137932.1 |
| chr5_-_59783882 | 0.13 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr3_-_62359180 | 0.13 |
ENST00000283268.3
|
FEZF2
|
FEZ family zinc finger 2 |
| chr11_-_8290263 | 0.13 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr11_-_8285405 | 0.13 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr1_-_102312517 | 0.13 |
ENST00000338858.5
|
OLFM3
|
olfactomedin 3 |
| chr14_-_69261310 | 0.12 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr3_-_114343039 | 0.12 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_+_6105974 | 0.12 |
ENST00000378083.3
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chrX_+_135252050 | 0.12 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr5_+_170736243 | 0.11 |
ENST00000296921.5
|
TLX3
|
T-cell leukemia homeobox 3 |
| chr17_-_39150385 | 0.11 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr7_-_31380502 | 0.11 |
ENST00000297142.3
|
NEUROD6
|
neuronal differentiation 6 |
| chr6_-_76782371 | 0.11 |
ENST00000369950.3
ENST00000369963.3 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr2_+_182850743 | 0.11 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chrX_-_19688475 | 0.11 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr17_-_46716647 | 0.10 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr1_-_116383322 | 0.10 |
ENST00000429731.1
|
NHLH2
|
nescient helix loop helix 2 |
| chr1_-_203055129 | 0.10 |
ENST00000241651.4
|
MYOG
|
myogenin (myogenic factor 4) |
| chr14_-_92413353 | 0.10 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr2_+_210444142 | 0.10 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr8_-_122653630 | 0.10 |
ENST00000303924.4
|
HAS2
|
hyaluronan synthase 2 |
| chr21_-_40033618 | 0.10 |
ENST00000417133.2
ENST00000398910.1 ENST00000442448.1 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr17_-_10421853 | 0.10 |
ENST00000226207.5
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult |
| chr7_-_5465045 | 0.10 |
ENST00000399434.2
|
TNRC18
|
trinucleotide repeat containing 18 |
| chr12_-_91398796 | 0.10 |
ENST00000261172.3
ENST00000551767.1 |
EPYC
|
epiphycan |
| chr6_-_22297730 | 0.09 |
ENST00000306482.1
|
PRL
|
prolactin |
| chr16_+_72459838 | 0.09 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chr6_-_10115007 | 0.09 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr10_+_24755416 | 0.09 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr6_-_85474219 | 0.09 |
ENST00000369663.5
|
TBX18
|
T-box 18 |
| chr2_+_54785485 | 0.09 |
ENST00000333896.5
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr1_+_204839959 | 0.09 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chr2_+_33359687 | 0.09 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr5_-_24645078 | 0.09 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr12_-_5352315 | 0.09 |
ENST00000536518.1
|
RP11-319E16.1
|
RP11-319E16.1 |
| chr15_+_58430567 | 0.09 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr18_+_59000815 | 0.09 |
ENST00000262717.4
|
CDH20
|
cadherin 20, type 2 |
| chr10_-_21786179 | 0.09 |
ENST00000377113.5
|
CASC10
|
cancer susceptibility candidate 10 |
| chr1_+_84630367 | 0.09 |
ENST00000370680.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr2_-_80531399 | 0.08 |
ENST00000409148.1
ENST00000415098.1 ENST00000452811.1 |
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr15_+_58724184 | 0.08 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr2_+_210444748 | 0.08 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr2_+_33359646 | 0.08 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr4_+_100495864 | 0.08 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr15_-_54025300 | 0.08 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr2_-_175711133 | 0.08 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr4_-_110223799 | 0.08 |
ENST00000399132.1
ENST00000399126.1 ENST00000505591.1 |
COL25A1
|
collagen, type XXV, alpha 1 |
| chr15_+_58430368 | 0.08 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr16_+_7382745 | 0.08 |
ENST00000436368.2
ENST00000311745.5 ENST00000355637.4 ENST00000340209.4 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr12_-_102591604 | 0.08 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr14_-_98444386 | 0.08 |
ENST00000556462.1
ENST00000556138.1 |
C14orf64
|
chromosome 14 open reading frame 64 |
| chr3_+_1134260 | 0.08 |
ENST00000446702.2
ENST00000539053.1 ENST00000350110.2 |
CNTN6
|
contactin 6 |
| chr12_-_14133053 | 0.08 |
ENST00000609686.1
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B |
| chr8_+_92261516 | 0.08 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr17_-_47045949 | 0.07 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr2_+_226265364 | 0.07 |
ENST00000272907.6
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr13_-_46425865 | 0.07 |
ENST00000400405.2
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr7_+_18535346 | 0.07 |
ENST00000405010.3
ENST00000406451.4 ENST00000428307.2 |
HDAC9
|
histone deacetylase 9 |
| chrX_-_15683147 | 0.07 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr7_-_14026123 | 0.07 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr4_+_41614720 | 0.07 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chrX_-_124097620 | 0.07 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr6_+_42584847 | 0.07 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr1_+_65730385 | 0.07 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr8_-_25281747 | 0.07 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr17_-_46671323 | 0.06 |
ENST00000239151.5
|
HOXB5
|
homeobox B5 |
| chr3_+_152017181 | 0.06 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr3_+_152017360 | 0.06 |
ENST00000485910.1
ENST00000463374.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr6_+_155537771 | 0.06 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr12_+_41831485 | 0.06 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr12_-_24103954 | 0.06 |
ENST00000441133.2
ENST00000545921.1 |
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr1_-_217250231 | 0.06 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr14_-_24551137 | 0.06 |
ENST00000396995.1
|
NRL
|
neural retina leucine zipper |
| chr13_-_84456527 | 0.06 |
ENST00000377084.2
|
SLITRK1
|
SLIT and NTRK-like family, member 1 |
| chr4_+_160188889 | 0.06 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr9_-_124989804 | 0.06 |
ENST00000373755.2
ENST00000373754.2 |
LHX6
|
LIM homeobox 6 |
| chr16_-_67517716 | 0.06 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr5_-_88179302 | 0.06 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr14_+_22977587 | 0.06 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr4_-_176733897 | 0.06 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr9_+_125796806 | 0.06 |
ENST00000373642.1
|
GPR21
|
G protein-coupled receptor 21 |
| chr2_+_171571827 | 0.06 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr10_-_106240032 | 0.06 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr14_-_24551195 | 0.06 |
ENST00000560550.1
|
NRL
|
neural retina leucine zipper |
| chr7_-_14026063 | 0.06 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr6_+_15401075 | 0.06 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr1_-_216896780 | 0.05 |
ENST00000459955.1
ENST00000366937.1 ENST00000408911.3 ENST00000391890.3 |
ESRRG
|
estrogen-related receptor gamma |
| chr5_+_140710061 | 0.05 |
ENST00000517417.1
ENST00000378105.3 |
PCDHGA1
|
protocadherin gamma subfamily A, 1 |
| chr15_-_70390191 | 0.05 |
ENST00000559191.1
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chrX_-_33146477 | 0.05 |
ENST00000378677.2
|
DMD
|
dystrophin |
| chr12_-_16761007 | 0.05 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr21_-_39870339 | 0.05 |
ENST00000429727.2
ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr2_-_233641265 | 0.05 |
ENST00000438786.1
ENST00000409779.1 ENST00000233826.3 |
KCNJ13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr2_-_145277569 | 0.05 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr6_-_10412600 | 0.05 |
ENST00000379608.3
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr4_+_71494461 | 0.05 |
ENST00000396073.3
|
ENAM
|
enamelin |
| chr2_-_77749474 | 0.05 |
ENST00000409093.1
ENST00000409088.3 |
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr10_+_11047259 | 0.05 |
ENST00000379261.4
ENST00000416382.2 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr11_+_20620946 | 0.05 |
ENST00000525748.1
|
SLC6A5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr15_+_69854027 | 0.05 |
ENST00000498938.2
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr14_-_57277163 | 0.05 |
ENST00000555006.1
|
OTX2
|
orthodenticle homeobox 2 |
| chr17_-_46688334 | 0.05 |
ENST00000239165.7
|
HOXB7
|
homeobox B7 |
| chr18_+_32073253 | 0.05 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr3_-_157824292 | 0.05 |
ENST00000483851.2
|
SHOX2
|
short stature homeobox 2 |
| chrX_+_28605516 | 0.05 |
ENST00000378993.1
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1 |
| chr5_+_63802109 | 0.05 |
ENST00000334025.2
|
RGS7BP
|
regulator of G-protein signaling 7 binding protein |
| chr3_+_137490748 | 0.05 |
ENST00000478772.1
|
RP11-2A4.3
|
RP11-2A4.3 |
| chr5_-_34043310 | 0.05 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr11_+_131240373 | 0.05 |
ENST00000374791.3
ENST00000436745.1 |
NTM
|
neurotrimin |
| chr12_-_117319236 | 0.05 |
ENST00000257572.5
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr12_+_81110684 | 0.04 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chr5_-_58295712 | 0.04 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr12_-_91576561 | 0.04 |
ENST00000547568.2
ENST00000552962.1 |
DCN
|
decorin |
| chr11_+_22696314 | 0.04 |
ENST00000532398.1
ENST00000433790.1 |
GAS2
|
growth arrest-specific 2 |
| chr3_-_57233966 | 0.04 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr1_+_62439037 | 0.04 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr7_-_112726393 | 0.04 |
ENST00000449591.1
ENST00000449735.1 ENST00000438062.1 ENST00000424100.1 |
GPR85
|
G protein-coupled receptor 85 |
| chr6_+_50681541 | 0.04 |
ENST00000008391.3
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr5_-_58882219 | 0.04 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr5_-_88119580 | 0.04 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr22_-_36236265 | 0.04 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr9_-_123812542 | 0.04 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr5_+_176811431 | 0.04 |
ENST00000512593.1
ENST00000324417.5 |
SLC34A1
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr1_+_41174988 | 0.04 |
ENST00000372652.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr17_-_46692287 | 0.04 |
ENST00000239144.4
|
HOXB8
|
homeobox B8 |
| chr1_+_186265399 | 0.04 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr12_-_87232644 | 0.04 |
ENST00000549405.2
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr14_+_97925151 | 0.04 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr21_-_31538971 | 0.04 |
ENST00000286808.3
|
CLDN17
|
claudin 17 |
| chr6_-_10415218 | 0.04 |
ENST00000466073.1
ENST00000498450.1 |
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr14_-_98444438 | 0.04 |
ENST00000512901.2
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr1_+_84630053 | 0.04 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr7_-_112727774 | 0.03 |
ENST00000297146.3
ENST00000501255.2 |
GPR85
|
G protein-coupled receptor 85 |
| chr17_-_46657473 | 0.03 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr7_-_5463175 | 0.03 |
ENST00000399537.4
ENST00000430969.1 |
TNRC18
|
trinucleotide repeat containing 18 |
| chr2_-_183291741 | 0.03 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr12_-_86650045 | 0.03 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr9_+_82186872 | 0.03 |
ENST00000376544.3
ENST00000376520.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr2_+_182850551 | 0.03 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr12_-_99548270 | 0.03 |
ENST00000546568.1
ENST00000332712.7 ENST00000546960.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr9_-_73483926 | 0.03 |
ENST00000396283.1
ENST00000361823.5 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr14_-_37051798 | 0.03 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr6_+_136172820 | 0.03 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr17_-_46608272 | 0.03 |
ENST00000577092.1
ENST00000239174.6 |
HOXB1
|
homeobox B1 |
| chr8_+_104892639 | 0.03 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr12_-_102874378 | 0.03 |
ENST00000456098.1
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr13_-_99667960 | 0.03 |
ENST00000448493.2
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr12_-_117318788 | 0.03 |
ENST00000550505.1
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr6_-_138820624 | 0.03 |
ENST00000343505.5
|
NHSL1
|
NHS-like 1 |
| chr12_-_102874416 | 0.03 |
ENST00000392904.1
ENST00000337514.6 |
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 0.7 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.3 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.2 | GO:0035283 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.2 | GO:0055018 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.3 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.3 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.0 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.1 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) optic cup structural organization(GO:0003409) |
| 0.0 | 0.2 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:1903862 | regulation of muscle atrophy(GO:0014735) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.0 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.0 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.0 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.0 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.0 | GO:0097475 | motor neuron migration(GO:0097475) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.2 | GO:0070700 | co-receptor binding(GO:0039706) BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.0 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |