Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
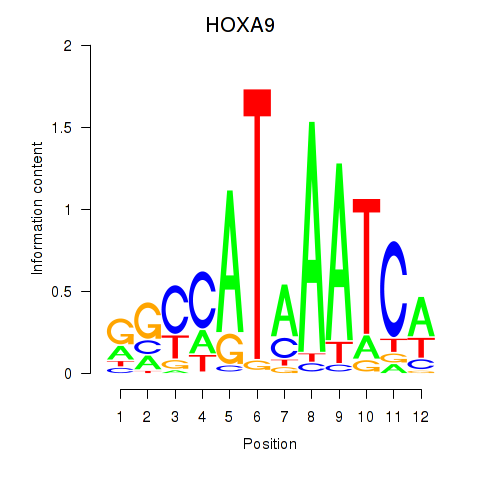
Results for HOXA9
Z-value: 0.61
Transcription factors associated with HOXA9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA9
|
ENSG00000078399.11 | homeobox A9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA9 | hg19_v2_chr7_-_27205136_27205164 | 0.15 | 4.4e-01 | Click! |
Activity profile of HOXA9 motif
Sorted Z-values of HOXA9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_55867146 | 1.93 |
ENST00000422046.2
|
CES1
|
carboxylesterase 1 |
| chr1_-_152131703 | 1.21 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr12_-_13248598 | 0.87 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr1_+_152956549 | 0.63 |
ENST00000307122.2
|
SPRR1A
|
small proline-rich protein 1A |
| chr2_-_88427568 | 0.57 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chr3_-_195310802 | 0.49 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr11_+_6866883 | 0.48 |
ENST00000299454.4
ENST00000379831.2 |
OR10A5
|
olfactory receptor, family 10, subfamily A, member 5 |
| chr1_+_24645807 | 0.47 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_+_24645865 | 0.46 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr9_-_127269661 | 0.44 |
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr6_-_27775694 | 0.43 |
ENST00000377401.2
|
HIST1H2BL
|
histone cluster 1, H2bl |
| chr2_+_190541153 | 0.43 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr3_+_186353756 | 0.41 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr2_+_89986318 | 0.41 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr4_-_150736962 | 0.40 |
ENST00000502345.1
ENST00000510975.1 ENST00000511993.1 |
RP11-526A4.1
|
RP11-526A4.1 |
| chr2_+_232063436 | 0.38 |
ENST00000440107.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr10_+_102222798 | 0.37 |
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B |
| chr6_-_46138676 | 0.37 |
ENST00000371383.2
ENST00000230565.3 |
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative) |
| chr16_-_67427389 | 0.36 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr4_-_156787425 | 0.36 |
ENST00000537611.2
|
ASIC5
|
acid-sensing (proton-gated) ion channel family member 5 |
| chrX_+_65384182 | 0.36 |
ENST00000441993.2
ENST00000419594.1 |
HEPH
|
hephaestin |
| chr4_-_34041504 | 0.35 |
ENST00000512581.1
ENST00000505018.1 |
RP11-79E3.3
|
RP11-79E3.3 |
| chr2_+_108994466 | 0.34 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr11_+_60048004 | 0.33 |
ENST00000532114.1
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr14_-_25479811 | 0.33 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr2_+_108994633 | 0.33 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr10_+_85933494 | 0.32 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr6_-_25832254 | 0.32 |
ENST00000476801.1
ENST00000244527.4 ENST00000427328.1 |
SLC17A1
|
solute carrier family 17 (organic anion transporter), member 1 |
| chr12_-_59175485 | 0.32 |
ENST00000550678.1
ENST00000552201.1 |
RP11-767I20.1
|
RP11-767I20.1 |
| chr12_-_52779433 | 0.32 |
ENST00000257951.3
|
KRT84
|
keratin 84 |
| chr3_-_149510553 | 0.31 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr3_+_148545586 | 0.30 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr7_-_3214287 | 0.30 |
ENST00000404626.3
|
AC091801.1
|
LOC392621; Uncharacterized protein |
| chr10_+_51549498 | 0.30 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chrX_-_110513703 | 0.29 |
ENST00000324068.1
|
CAPN6
|
calpain 6 |
| chr2_-_197226875 | 0.29 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr11_-_59952106 | 0.28 |
ENST00000529054.1
ENST00000530839.1 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr6_-_138539627 | 0.28 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr22_+_23248512 | 0.28 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr2_+_11864458 | 0.27 |
ENST00000396098.1
ENST00000396099.1 ENST00000425416.2 |
LPIN1
|
lipin 1 |
| chrX_+_144908928 | 0.27 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr7_-_27169801 | 0.27 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr2_-_209010874 | 0.26 |
ENST00000260988.4
|
CRYGB
|
crystallin, gamma B |
| chr11_-_59951889 | 0.26 |
ENST00000532169.1
ENST00000534596.1 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr22_+_30477000 | 0.26 |
ENST00000403975.1
|
HORMAD2
|
HORMA domain containing 2 |
| chr2_+_20646824 | 0.25 |
ENST00000272233.4
|
RHOB
|
ras homolog family member B |
| chr1_+_24646002 | 0.25 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr3_-_108836977 | 0.25 |
ENST00000232603.5
|
MORC1
|
MORC family CW-type zinc finger 1 |
| chr5_-_101834617 | 0.25 |
ENST00000513675.1
ENST00000379807.3 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chr22_-_30642782 | 0.24 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr21_-_19775973 | 0.24 |
ENST00000284885.3
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr14_+_88471468 | 0.24 |
ENST00000267549.3
|
GPR65
|
G protein-coupled receptor 65 |
| chr3_-_197686847 | 0.24 |
ENST00000265239.6
|
IQCG
|
IQ motif containing G |
| chr8_+_38261880 | 0.24 |
ENST00000527175.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chrX_-_45629661 | 0.24 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr3_+_178276488 | 0.24 |
ENST00000432997.1
ENST00000455865.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr5_-_101834712 | 0.24 |
ENST00000506729.1
ENST00000389019.3 ENST00000379810.1 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chr5_-_33297946 | 0.24 |
ENST00000510327.1
|
CTD-2066L21.3
|
CTD-2066L21.3 |
| chr15_-_31283618 | 0.23 |
ENST00000563714.1
|
MTMR10
|
myotubularin related protein 10 |
| chr9_+_78505554 | 0.23 |
ENST00000545128.1
|
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr6_-_29399744 | 0.23 |
ENST00000377154.1
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1 |
| chr17_-_29624343 | 0.23 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr19_-_52598958 | 0.23 |
ENST00000594440.1
ENST00000426391.2 ENST00000389534.4 |
ZNF841
|
zinc finger protein 841 |
| chr1_-_43282906 | 0.22 |
ENST00000372521.4
|
CCDC23
|
coiled-coil domain containing 23 |
| chr10_+_82009466 | 0.22 |
ENST00000356374.4
|
AL359195.1
|
Uncharacterized protein; cDNA FLJ46261 fis, clone TESTI4025062 |
| chrX_+_65382433 | 0.21 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr13_+_52586517 | 0.21 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr5_-_94417339 | 0.21 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chrX_+_95939711 | 0.21 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr10_+_91092241 | 0.20 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr7_+_21582638 | 0.20 |
ENST00000409508.3
ENST00000328843.6 |
DNAH11
|
dynein, axonemal, heavy chain 11 |
| chr1_+_109256067 | 0.20 |
ENST00000271311.2
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr2_+_27799389 | 0.20 |
ENST00000408964.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chr16_+_70613770 | 0.20 |
ENST00000429149.2
ENST00000563721.2 |
IL34
|
interleukin 34 |
| chr1_-_205819245 | 0.19 |
ENST00000367136.4
|
PM20D1
|
peptidase M20 domain containing 1 |
| chrX_+_65384052 | 0.19 |
ENST00000336279.5
ENST00000458621.1 |
HEPH
|
hephaestin |
| chr7_-_16505440 | 0.19 |
ENST00000307068.4
|
SOSTDC1
|
sclerostin domain containing 1 |
| chr5_-_131347306 | 0.19 |
ENST00000296869.4
ENST00000379249.3 ENST00000379272.2 ENST00000379264.2 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr4_+_110834033 | 0.19 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chr7_+_54609995 | 0.19 |
ENST00000302287.3
ENST00000407838.3 |
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr2_+_138722028 | 0.19 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chrX_+_130192318 | 0.19 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr15_+_40697988 | 0.19 |
ENST00000487418.2
ENST00000479013.2 |
IVD
|
isovaleryl-CoA dehydrogenase |
| chr18_+_3466248 | 0.19 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr1_-_43282945 | 0.18 |
ENST00000537227.1
|
CCDC23
|
coiled-coil domain containing 23 |
| chr9_-_21228221 | 0.18 |
ENST00000413767.2
|
IFNA17
|
interferon, alpha 17 |
| chr15_+_40643227 | 0.18 |
ENST00000448599.2
|
PHGR1
|
proline/histidine/glycine-rich 1 |
| chr7_+_69064566 | 0.18 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr3_+_157154578 | 0.18 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr3_-_196911002 | 0.18 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr11_-_76155618 | 0.18 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr17_-_8021710 | 0.18 |
ENST00000380149.1
ENST00000448843.2 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr12_+_21207503 | 0.17 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr4_+_48018781 | 0.17 |
ENST00000295461.5
|
NIPAL1
|
NIPA-like domain containing 1 |
| chr1_-_152297679 | 0.17 |
ENST00000368799.1
|
FLG
|
filaggrin |
| chr11_+_57310114 | 0.17 |
ENST00000527972.1
ENST00000399154.2 |
SMTNL1
|
smoothelin-like 1 |
| chr6_+_35310312 | 0.17 |
ENST00000448077.2
ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr16_+_29690358 | 0.17 |
ENST00000395384.4
ENST00000562473.1 |
QPRT
|
quinolinate phosphoribosyltransferase |
| chr8_+_76452097 | 0.16 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr11_+_71927807 | 0.16 |
ENST00000298223.6
ENST00000454954.2 ENST00000541003.1 ENST00000539412.1 ENST00000536778.1 ENST00000535625.1 ENST00000321324.7 |
FOLR2
|
folate receptor 2 (fetal) |
| chr22_-_31364187 | 0.16 |
ENST00000215862.4
ENST00000397641.3 |
MORC2
|
MORC family CW-type zinc finger 2 |
| chr18_-_72264805 | 0.16 |
ENST00000577806.1
|
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chr5_-_94417314 | 0.16 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr2_-_225811747 | 0.16 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr19_-_50529193 | 0.16 |
ENST00000596445.1
ENST00000599538.1 |
VRK3
|
vaccinia related kinase 3 |
| chr5_+_149877334 | 0.16 |
ENST00000523767.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr16_+_16434185 | 0.16 |
ENST00000524823.2
|
AC138969.4
|
Protein PKD1P1 |
| chr15_+_63414760 | 0.15 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr3_-_190167571 | 0.15 |
ENST00000354905.2
|
TMEM207
|
transmembrane protein 207 |
| chr1_-_7913089 | 0.15 |
ENST00000361696.5
|
UTS2
|
urotensin 2 |
| chr9_+_78505581 | 0.15 |
ENST00000376767.3
ENST00000376752.4 |
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr21_-_46012386 | 0.15 |
ENST00000400368.1
|
KRTAP10-6
|
keratin associated protein 10-6 |
| chr3_+_108321623 | 0.15 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr11_+_59522837 | 0.15 |
ENST00000437946.2
|
STX3
|
syntaxin 3 |
| chr12_-_121477039 | 0.15 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr10_-_14050522 | 0.15 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr22_-_31324215 | 0.15 |
ENST00000429468.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr7_+_20370746 | 0.15 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr10_-_45474237 | 0.15 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chr6_-_133035185 | 0.15 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr16_-_18462221 | 0.15 |
ENST00000528301.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr2_-_130031335 | 0.14 |
ENST00000375987.3
|
AC079586.1
|
AC079586.1 |
| chr5_+_126984710 | 0.14 |
ENST00000379445.3
|
CTXN3
|
cortexin 3 |
| chr4_-_76439483 | 0.14 |
ENST00000380840.2
ENST00000513257.1 ENST00000507014.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr15_+_59730348 | 0.14 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr2_+_138721850 | 0.14 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr15_-_68497657 | 0.14 |
ENST00000448060.2
ENST00000467889.1 |
CALML4
|
calmodulin-like 4 |
| chr12_-_91573316 | 0.14 |
ENST00000393155.1
|
DCN
|
decorin |
| chr10_+_47894572 | 0.14 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr18_-_72265035 | 0.14 |
ENST00000585279.1
ENST00000580048.1 |
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chr1_+_101185290 | 0.14 |
ENST00000370119.4
ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr1_-_85100703 | 0.13 |
ENST00000370624.1
|
C1orf180
|
chromosome 1 open reading frame 180 |
| chr12_-_121476959 | 0.13 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chrX_+_95939638 | 0.13 |
ENST00000373061.3
ENST00000373054.4 ENST00000355827.4 |
DIAPH2
|
diaphanous-related formin 2 |
| chr8_+_20831460 | 0.13 |
ENST00000522604.1
|
RP11-421P23.1
|
RP11-421P23.1 |
| chr4_-_74964904 | 0.13 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr15_+_65134088 | 0.13 |
ENST00000323544.4
ENST00000437723.1 |
PLEKHO2
AC069368.3
|
pleckstrin homology domain containing, family O member 2 Uncharacterized protein |
| chrX_+_135388147 | 0.13 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr6_+_24126350 | 0.12 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr10_-_96829246 | 0.12 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr5_+_175490540 | 0.12 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr1_+_50575292 | 0.12 |
ENST00000371821.1
ENST00000371819.1 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr14_+_78870030 | 0.12 |
ENST00000553631.1
ENST00000554719.1 |
NRXN3
|
neurexin 3 |
| chr4_-_110723134 | 0.12 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr3_-_151176497 | 0.12 |
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr3_+_26664291 | 0.12 |
ENST00000396641.2
|
LRRC3B
|
leucine rich repeat containing 3B |
| chr8_+_31497271 | 0.12 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr11_-_35441597 | 0.12 |
ENST00000395753.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr17_+_57297807 | 0.12 |
ENST00000284116.4
ENST00000581140.1 ENST00000581276.1 |
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr11_-_108464465 | 0.12 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr16_-_21868739 | 0.12 |
ENST00000415645.2
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr13_-_30881134 | 0.12 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr11_-_108464321 | 0.12 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chr7_+_128312346 | 0.12 |
ENST00000480462.1
ENST00000378704.3 ENST00000477515.1 |
FAM71F2
|
family with sequence similarity 71, member F2 |
| chr1_+_101702417 | 0.11 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr2_+_182850551 | 0.11 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr12_-_71148413 | 0.11 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr4_-_76944621 | 0.11 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chrX_+_30248553 | 0.11 |
ENST00000361644.2
|
MAGEB3
|
melanoma antigen family B, 3 |
| chr17_+_7758374 | 0.11 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr6_+_35310391 | 0.11 |
ENST00000337400.2
ENST00000311565.4 ENST00000540939.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr1_-_38512450 | 0.11 |
ENST00000373012.2
|
POU3F1
|
POU class 3 homeobox 1 |
| chr1_-_153085984 | 0.11 |
ENST00000468739.1
|
SPRR2F
|
small proline-rich protein 2F |
| chr5_+_111755280 | 0.11 |
ENST00000600409.1
|
EPB41L4A-AS2
|
EPB41L4A antisense RNA 2 (head to head) |
| chr11_-_5345582 | 0.11 |
ENST00000328813.2
|
OR51B2
|
olfactory receptor, family 51, subfamily B, member 2 |
| chr21_+_44313375 | 0.11 |
ENST00000354250.2
ENST00000340344.4 |
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr3_+_120626919 | 0.11 |
ENST00000273666.6
ENST00000471454.1 ENST00000472879.1 ENST00000497029.1 ENST00000492541.1 |
STXBP5L
|
syntaxin binding protein 5-like |
| chr17_+_44370099 | 0.11 |
ENST00000496930.1
|
LRRC37A
|
leucine rich repeat containing 37A |
| chr15_+_94899183 | 0.11 |
ENST00000557742.1
|
MCTP2
|
multiple C2 domains, transmembrane 2 |
| chr16_-_20338748 | 0.11 |
ENST00000575582.1
ENST00000341642.5 ENST00000381362.4 ENST00000572347.1 ENST00000572478.1 ENST00000302555.5 |
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr18_+_32558208 | 0.10 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr15_+_67418047 | 0.10 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr16_-_52119019 | 0.10 |
ENST00000561513.1
ENST00000565742.1 |
LINC00919
|
long intergenic non-protein coding RNA 919 |
| chr4_-_110723194 | 0.10 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr8_+_145215928 | 0.10 |
ENST00000528919.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr15_+_33022885 | 0.10 |
ENST00000322805.4
|
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr14_+_22977587 | 0.10 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr19_+_37341752 | 0.10 |
ENST00000586933.1
ENST00000532141.1 ENST00000420450.1 ENST00000526123.1 |
ZNF345
|
zinc finger protein 345 |
| chr6_+_126240442 | 0.10 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr1_+_226250379 | 0.10 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chr21_+_37507210 | 0.10 |
ENST00000290354.5
|
CBR3
|
carbonyl reductase 3 |
| chr9_-_21207142 | 0.10 |
ENST00000357374.2
|
IFNA10
|
interferon, alpha 10 |
| chr7_-_80141328 | 0.10 |
ENST00000398291.3
|
GNAT3
|
guanine nucleotide binding protein, alpha transducing 3 |
| chr1_+_44584522 | 0.10 |
ENST00000372299.3
|
KLF17
|
Kruppel-like factor 17 |
| chr19_-_42133420 | 0.10 |
ENST00000221954.2
ENST00000600925.1 |
CEACAM4
|
carcinoembryonic antigen-related cell adhesion molecule 4 |
| chr7_-_92855762 | 0.10 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr11_-_3400330 | 0.10 |
ENST00000427810.2
ENST00000005082.9 ENST00000534569.1 ENST00000438262.2 ENST00000528796.1 ENST00000528410.1 ENST00000529678.1 ENST00000354599.6 ENST00000526601.1 ENST00000525502.1 ENST00000533036.1 ENST00000399602.4 |
ZNF195
|
zinc finger protein 195 |
| chr1_-_36948879 | 0.10 |
ENST00000373106.1
ENST00000373104.1 ENST00000373103.1 |
CSF3R
|
colony stimulating factor 3 receptor (granulocyte) |
| chr8_-_25281747 | 0.09 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr10_+_24738355 | 0.09 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr17_-_7232585 | 0.09 |
ENST00000571887.1
ENST00000315614.7 ENST00000399464.2 ENST00000570460.1 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr15_-_41166414 | 0.09 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr14_-_24911448 | 0.09 |
ENST00000555355.1
ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr7_+_44084233 | 0.09 |
ENST00000448521.1
|
DBNL
|
drebrin-like |
| chr11_-_76155700 | 0.09 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr3_-_72149553 | 0.09 |
ENST00000468646.2
ENST00000464271.1 |
LINC00877
|
long intergenic non-protein coding RNA 877 |
| chr19_-_47975417 | 0.09 |
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr11_-_126870655 | 0.09 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr22_+_23077065 | 0.09 |
ENST00000390310.2
|
IGLV2-18
|
immunoglobulin lambda variable 2-18 |
| chr13_+_77522632 | 0.09 |
ENST00000377462.1
|
IRG1
|
immunoresponsive 1 homolog (mouse) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.2 | 0.5 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.4 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.4 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 1.2 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.6 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.7 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 0.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.2 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.2 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.2 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.2 | GO:0071231 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.3 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.4 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.3 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.2 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.2 | GO:1903764 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.2 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.3 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 | 0.3 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.2 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) regulation of neurotrophin production(GO:0032899) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 1.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:2000400 | pantothenate metabolic process(GO:0015939) positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.0 | 0.2 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0032916 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 | 0.0 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.0 | 0.0 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.0 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 2.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.0 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 0.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.2 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.2 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.2 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.4 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |