Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
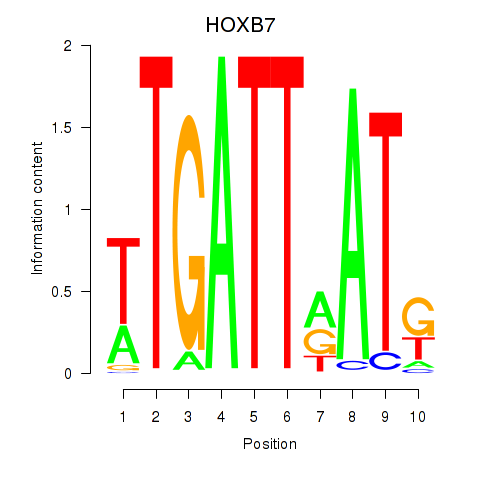
Results for HOXB7
Z-value: 0.65
Transcription factors associated with HOXB7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB7
|
ENSG00000260027.3 | homeobox B7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB7 | hg19_v2_chr17_-_46688334_46688385 | 0.14 | 4.5e-01 | Click! |
Activity profile of HOXB7 motif
Sorted Z-values of HOXB7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_91095099 | 1.33 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr9_+_105757590 | 1.02 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr21_+_41029235 | 1.01 |
ENST00000380618.1
|
B3GALT5
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 |
| chr4_+_69962212 | 0.97 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr4_+_69962185 | 0.96 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr1_+_152956549 | 0.85 |
ENST00000307122.2
|
SPRR1A
|
small proline-rich protein 1A |
| chr19_-_14889349 | 0.82 |
ENST00000315576.3
ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr12_+_101988774 | 0.81 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr12_+_20968608 | 0.81 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr13_-_103719196 | 0.80 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr12_+_101988627 | 0.80 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr12_+_21168630 | 0.78 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr8_-_7638935 | 0.75 |
ENST00000528972.1
|
AC084121.16
|
proline rich 23 domain containing 2 |
| chr1_-_152131703 | 0.74 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr9_-_93405352 | 0.71 |
ENST00000375765.3
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr2_+_234104079 | 0.70 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr16_-_46655538 | 0.69 |
ENST00000303383.3
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr5_+_140227357 | 0.69 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr4_+_69681710 | 0.67 |
ENST00000265403.7
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr8_+_7397150 | 0.67 |
ENST00000533250.1
|
RP11-1118M6.1
|
proline rich 23 domain containing 1 |
| chr2_-_190044480 | 0.63 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr4_-_70080449 | 0.63 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr11_+_57365150 | 0.62 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr12_-_28124903 | 0.59 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr10_+_102891048 | 0.57 |
ENST00000467928.2
|
TLX1
|
T-cell leukemia homeobox 1 |
| chr11_-_107729887 | 0.56 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr4_-_48014931 | 0.55 |
ENST00000420489.2
ENST00000504722.1 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr1_+_101702417 | 0.55 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr12_-_22063787 | 0.54 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr10_+_124320195 | 0.53 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr4_+_70146217 | 0.53 |
ENST00000335568.5
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chr9_-_32526299 | 0.51 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr14_+_35591858 | 0.51 |
ENST00000603544.1
|
KIAA0391
|
KIAA0391 |
| chr4_+_169013666 | 0.49 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr3_+_189349162 | 0.49 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr8_-_117043 | 0.49 |
ENST00000320901.3
|
OR4F21
|
olfactory receptor, family 4, subfamily F, member 21 |
| chr10_+_124320156 | 0.49 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr12_-_91573132 | 0.48 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr2_+_27719697 | 0.48 |
ENST00000264717.2
ENST00000424318.2 |
GCKR
|
glucokinase (hexokinase 4) regulator |
| chr12_-_28125638 | 0.47 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr11_-_2162162 | 0.46 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr12_-_95510743 | 0.46 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr11_-_67981046 | 0.46 |
ENST00000402789.1
ENST00000402185.2 ENST00000458496.1 |
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr4_+_71200681 | 0.45 |
ENST00000273936.5
|
CABS1
|
calcium-binding protein, spermatid-specific 1 |
| chr1_-_200589859 | 0.45 |
ENST00000367350.4
|
KIF14
|
kinesin family member 14 |
| chr3_+_101818088 | 0.45 |
ENST00000491959.1
|
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr5_-_179047881 | 0.43 |
ENST00000521173.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr2_-_70780770 | 0.42 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr6_-_66417107 | 0.41 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr18_+_22040620 | 0.41 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr6_+_72926145 | 0.41 |
ENST00000425662.2
ENST00000453976.2 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr2_+_89986318 | 0.40 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr11_+_844406 | 0.40 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr13_+_57721622 | 0.39 |
ENST00000377930.1
|
PRR20B
|
proline rich 20B |
| chr3_-_58613323 | 0.38 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr19_-_51512804 | 0.38 |
ENST00000594211.1
ENST00000376832.4 |
KLK9
|
kallikrein-related peptidase 9 |
| chr17_-_64225508 | 0.38 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr8_-_110986918 | 0.38 |
ENST00000297404.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr5_-_179045199 | 0.37 |
ENST00000523921.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr16_-_88729473 | 0.37 |
ENST00000301012.3
ENST00000569177.1 |
MVD
|
mevalonate (diphospho) decarboxylase |
| chr5_+_162887556 | 0.37 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr12_-_91573316 | 0.36 |
ENST00000393155.1
|
DCN
|
decorin |
| chr8_+_39770803 | 0.36 |
ENST00000518237.1
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr11_+_44117099 | 0.36 |
ENST00000533608.1
|
EXT2
|
exostosin glycosyltransferase 2 |
| chr6_+_130339710 | 0.35 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr1_+_367640 | 0.35 |
ENST00000426406.1
|
OR4F29
|
olfactory receptor, family 4, subfamily F, member 29 |
| chr5_-_76788024 | 0.35 |
ENST00000515253.1
ENST00000414719.2 ENST00000507654.1 ENST00000514559.1 ENST00000511791.1 |
WDR41
|
WD repeat domain 41 |
| chr18_+_22040593 | 0.35 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr7_-_86595190 | 0.34 |
ENST00000398276.2
ENST00000416314.1 ENST00000425689.1 |
KIAA1324L
|
KIAA1324-like |
| chr9_+_125132803 | 0.34 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr5_-_171881362 | 0.34 |
ENST00000519643.1
|
SH3PXD2B
|
SH3 and PX domains 2B |
| chr7_+_26332645 | 0.34 |
ENST00000396376.1
|
SNX10
|
sorting nexin 10 |
| chr14_+_51955831 | 0.34 |
ENST00000356218.4
|
FRMD6
|
FERM domain containing 6 |
| chr5_-_127418573 | 0.33 |
ENST00000508353.1
ENST00000508878.1 ENST00000501652.1 ENST00000514409.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr4_+_148653206 | 0.33 |
ENST00000336498.3
|
ARHGAP10
|
Rho GTPase activating protein 10 |
| chr20_-_50179368 | 0.32 |
ENST00000609943.1
ENST00000609507.1 |
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
| chr1_-_622053 | 0.32 |
ENST00000332831.2
|
OR4F16
|
olfactory receptor, family 4, subfamily F, member 16 |
| chr16_-_30122717 | 0.32 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr7_-_155604967 | 0.31 |
ENST00000297261.2
|
SHH
|
sonic hedgehog |
| chr3_-_42452050 | 0.31 |
ENST00000441172.1
ENST00000287748.3 |
LYZL4
|
lysozyme-like 4 |
| chr18_+_29171689 | 0.31 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr12_-_91546926 | 0.31 |
ENST00000550758.1
|
DCN
|
decorin |
| chr3_-_141747950 | 0.31 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr3_+_52570610 | 0.31 |
ENST00000307106.3
ENST00000477703.1 ENST00000476842.1 |
SMIM4
|
small integral membrane protein 4 |
| chr1_-_67142710 | 0.30 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr11_+_113779704 | 0.30 |
ENST00000537778.1
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr20_-_1309809 | 0.30 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr2_-_228028829 | 0.30 |
ENST00000396625.3
ENST00000329662.7 |
COL4A4
|
collagen, type IV, alpha 4 |
| chr13_+_57728195 | 0.29 |
ENST00000544357.2
|
PRR20C
|
proline rich 20C |
| chr13_+_57734766 | 0.29 |
ENST00000452123.3
|
PRR20D
|
proline rich 20D |
| chr13_+_57741331 | 0.29 |
ENST00000434815.1
|
PRR20E
|
proline rich 20E |
| chr11_+_44117260 | 0.29 |
ENST00000358681.4
|
EXT2
|
exostosin glycosyltransferase 2 |
| chr15_+_67418047 | 0.29 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr7_-_36764142 | 0.29 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr9_-_73029540 | 0.29 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr3_+_189507432 | 0.28 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr14_+_74417192 | 0.28 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6 monooxygenase |
| chr6_+_34204642 | 0.28 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr17_-_41739283 | 0.28 |
ENST00000393661.2
ENST00000318579.4 |
MEOX1
|
mesenchyme homeobox 1 |
| chr9_+_125133315 | 0.28 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr5_+_131409476 | 0.27 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chrX_+_120181457 | 0.27 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr1_-_85100703 | 0.27 |
ENST00000370624.1
|
C1orf180
|
chromosome 1 open reading frame 180 |
| chr9_-_127269661 | 0.27 |
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr14_-_23624511 | 0.27 |
ENST00000529705.2
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr12_+_52056548 | 0.27 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr3_+_189507523 | 0.27 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr8_-_130799134 | 0.26 |
ENST00000276708.4
|
GSDMC
|
gasdermin C |
| chr10_+_13141585 | 0.26 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr17_-_41738931 | 0.26 |
ENST00000329168.3
ENST00000549132.1 |
MEOX1
|
mesenchyme homeobox 1 |
| chr3_+_156009623 | 0.26 |
ENST00000389634.5
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr9_-_73736511 | 0.26 |
ENST00000377110.3
ENST00000377111.2 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr14_+_55034599 | 0.26 |
ENST00000392067.3
ENST00000357634.3 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr3_-_24207039 | 0.26 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr12_-_54653313 | 0.25 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr4_+_187187098 | 0.25 |
ENST00000403665.2
ENST00000264692.4 |
F11
|
coagulation factor XI |
| chr10_+_13142075 | 0.25 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr8_+_31497271 | 0.25 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr11_-_57194948 | 0.25 |
ENST00000533235.1
ENST00000526621.1 ENST00000352187.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr2_+_176972000 | 0.24 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chr18_+_61554932 | 0.24 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr14_-_106830057 | 0.24 |
ENST00000390616.2
|
IGHV4-34
|
immunoglobulin heavy variable 4-34 |
| chr5_-_82969405 | 0.24 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr1_-_209957882 | 0.24 |
ENST00000294811.1
|
C1orf74
|
chromosome 1 open reading frame 74 |
| chr7_+_117824086 | 0.24 |
ENST00000249299.2
ENST00000424702.1 |
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr2_+_234637754 | 0.24 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr17_-_57229155 | 0.24 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr4_-_77328458 | 0.24 |
ENST00000388914.3
ENST00000434846.2 |
CCDC158
|
coiled-coil domain containing 158 |
| chr5_+_57787254 | 0.23 |
ENST00000502276.1
ENST00000396776.2 ENST00000511930.1 |
GAPT
|
GRB2-binding adaptor protein, transmembrane |
| chr8_+_9953214 | 0.23 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chr14_-_101295407 | 0.23 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr7_+_120629653 | 0.23 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr1_+_119957554 | 0.23 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr2_+_234621551 | 0.23 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr4_+_38511367 | 0.23 |
ENST00000507056.1
|
RP11-213G21.1
|
RP11-213G21.1 |
| chr2_+_105050794 | 0.23 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr12_-_7596735 | 0.23 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chrX_+_1387693 | 0.22 |
ENST00000381529.3
ENST00000432318.2 ENST00000361536.3 ENST00000501036.2 ENST00000381524.3 ENST00000412290.1 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr7_+_117824210 | 0.22 |
ENST00000422760.1
ENST00000411938.1 |
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr10_+_13141441 | 0.22 |
ENST00000263036.5
|
OPTN
|
optineurin |
| chr2_+_234826016 | 0.22 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr8_+_9953061 | 0.22 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr4_-_120243545 | 0.22 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr10_+_118305435 | 0.22 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr9_+_124103625 | 0.21 |
ENST00000594963.1
|
AL161784.1
|
Uncharacterized protein |
| chr12_-_86650077 | 0.21 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr2_+_171036635 | 0.21 |
ENST00000484338.2
ENST00000334231.6 |
MYO3B
|
myosin IIIB |
| chr15_+_58430567 | 0.21 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr9_+_128509624 | 0.21 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr10_+_5488564 | 0.21 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr21_+_43619796 | 0.21 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr6_-_137539651 | 0.21 |
ENST00000543628.1
|
IFNGR1
|
interferon gamma receptor 1 |
| chr15_+_58430368 | 0.21 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr1_+_117963209 | 0.21 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr2_+_102413726 | 0.21 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr12_+_56324933 | 0.21 |
ENST00000549629.1
ENST00000555218.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr11_-_104893863 | 0.20 |
ENST00000260315.3
ENST00000526056.1 ENST00000531367.1 ENST00000456094.1 ENST00000444749.2 ENST00000393141.2 ENST00000418434.1 ENST00000393139.2 |
CASP5
|
caspase 5, apoptosis-related cysteine peptidase |
| chrX_+_1387721 | 0.20 |
ENST00000419094.1
ENST00000381509.3 ENST00000494969.2 ENST00000355805.2 ENST00000355432.3 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr10_+_13142225 | 0.20 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr15_-_23692381 | 0.20 |
ENST00000567107.1
ENST00000345070.5 ENST00000312015.5 |
GOLGA6L2
|
golgin A6 family-like 2 |
| chr2_-_109605663 | 0.20 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr10_+_48189612 | 0.20 |
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr3_-_142166846 | 0.20 |
ENST00000463916.1
ENST00000544157.1 |
XRN1
|
5'-3' exoribonuclease 1 |
| chr6_+_167704838 | 0.20 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr8_+_41386761 | 0.20 |
ENST00000523277.2
|
GINS4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr16_+_66442411 | 0.20 |
ENST00000499966.1
|
LINC00920
|
long intergenic non-protein coding RNA 920 |
| chr9_-_27529726 | 0.20 |
ENST00000262244.5
|
MOB3B
|
MOB kinase activator 3B |
| chr10_+_69865866 | 0.19 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr15_-_28957469 | 0.19 |
ENST00000563027.1
ENST00000340249.3 |
GOLGA8M
|
golgin A8 family, member M |
| chr12_+_8309630 | 0.19 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr2_-_188378368 | 0.19 |
ENST00000392365.1
ENST00000435414.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr8_+_12803176 | 0.19 |
ENST00000524591.2
|
KIAA1456
|
KIAA1456 |
| chr8_+_104892639 | 0.19 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr10_+_50887683 | 0.19 |
ENST00000374113.3
ENST00000374111.3 ENST00000374112.3 ENST00000535836.1 |
C10orf53
|
chromosome 10 open reading frame 53 |
| chr18_+_52495426 | 0.18 |
ENST00000262094.5
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr9_-_128246769 | 0.18 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr19_+_54369608 | 0.18 |
ENST00000336967.3
|
MYADM
|
myeloid-associated differentiation marker |
| chrX_+_30248553 | 0.18 |
ENST00000361644.2
|
MAGEB3
|
melanoma antigen family B, 3 |
| chr19_-_33360647 | 0.18 |
ENST00000590341.1
ENST00000587772.1 ENST00000023064.4 |
SLC7A9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr1_-_241520525 | 0.18 |
ENST00000366565.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr19_-_52598958 | 0.18 |
ENST00000594440.1
ENST00000426391.2 ENST00000389534.4 |
ZNF841
|
zinc finger protein 841 |
| chr2_+_192141611 | 0.18 |
ENST00000392316.1
|
MYO1B
|
myosin IB |
| chr6_+_114178512 | 0.17 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr3_-_190167571 | 0.17 |
ENST00000354905.2
|
TMEM207
|
transmembrane protein 207 |
| chr2_-_99917639 | 0.17 |
ENST00000308528.4
|
LYG1
|
lysozyme G-like 1 |
| chrX_+_2976652 | 0.17 |
ENST00000537104.1
|
ARSF
|
arylsulfatase F |
| chr1_+_112016414 | 0.17 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr14_-_34931458 | 0.17 |
ENST00000298130.4
|
SPTSSA
|
serine palmitoyltransferase, small subunit A |
| chr1_+_158223923 | 0.17 |
ENST00000289429.5
|
CD1A
|
CD1a molecule |
| chr20_+_25388293 | 0.17 |
ENST00000262460.4
ENST00000429262.2 |
GINS1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr12_+_96252706 | 0.17 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr16_+_12059091 | 0.17 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr4_-_118006697 | 0.17 |
ENST00000310754.4
|
TRAM1L1
|
translocation associated membrane protein 1-like 1 |
| chr9_-_101471479 | 0.17 |
ENST00000259455.2
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr15_-_32695396 | 0.16 |
ENST00000512626.2
ENST00000435655.2 |
GOLGA8K
AC139426.2
|
golgin A8 family, member K Uncharacterized protein; cDNA FLJ52611 |
| chr6_+_54172653 | 0.16 |
ENST00000370869.3
|
TINAG
|
tubulointerstitial nephritis antigen |
| chr14_-_24584138 | 0.16 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chrX_+_13707235 | 0.16 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr6_+_27925019 | 0.16 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr2_+_234580525 | 0.16 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.2 | 0.7 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.2 | 0.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.5 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.1 | 1.3 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 0.9 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.1 | 0.9 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.5 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 2.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.4 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.9 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.4 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.4 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 1.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.3 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.1 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 0.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.5 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 1.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.2 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.1 | 0.3 | GO:2000195 | primary sex determination(GO:0007538) negative regulation of female gonad development(GO:2000195) |
| 0.1 | 0.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 1.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.3 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.5 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.1 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.1 | 0.3 | GO:0045918 | positive regulation of interleukin-23 production(GO:0032747) negative regulation of cytolysis(GO:0045918) |
| 0.1 | 2.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.2 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.0 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0035283 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.3 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.5 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.2 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.9 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.6 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.9 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.2 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.4 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 1.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.2 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.2 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.0 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.0 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.0 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.2 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:1903799 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.3 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.3 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.0 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.0 | GO:0050821 | protein stabilization(GO:0050821) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.2 | 0.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.5 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.5 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.6 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.5 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 1.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 1.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.2 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 1.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.0 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 0.5 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.2 | 0.6 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.2 | 0.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.6 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 4.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.4 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 0.7 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.4 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.5 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.3 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.5 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 1.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 1.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.0 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.2 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.5 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 1.0 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 1.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.0 | GO:0047023 | enone reductase activity(GO:0035671) androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.5 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 1.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.7 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |