Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
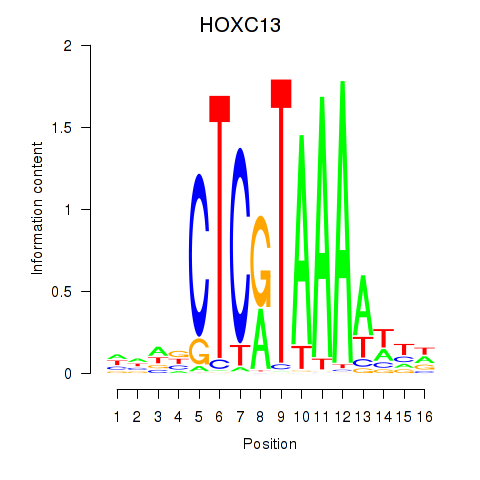
Results for HOXC13
Z-value: 0.79
Transcription factors associated with HOXC13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC13
|
ENSG00000123364.3 | homeobox C13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC13 | hg19_v2_chr12_+_54332535_54332636 | -0.56 | 1.5e-03 | Click! |
Activity profile of HOXC13 motif
Sorted Z-values of HOXC13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_88117683 | 2.41 |
ENST00000369562.4
|
C6ORF165
|
UPF0704 protein C6orf165 |
| chr1_-_47407097 | 2.04 |
ENST00000457840.2
|
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr1_-_109656439 | 1.96 |
ENST00000369949.4
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr13_-_20077417 | 1.75 |
ENST00000382978.1
ENST00000400230.2 ENST00000255310.6 |
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr15_+_71228826 | 1.59 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr10_+_51549498 | 1.53 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr4_+_70861647 | 1.19 |
ENST00000246895.4
ENST00000381060.2 |
STATH
|
statherin |
| chr15_+_71184931 | 1.16 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr4_-_147866960 | 1.15 |
ENST00000513335.1
|
TTC29
|
tetratricopeptide repeat domain 29 |
| chr15_+_71185148 | 1.08 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr17_-_39165366 | 1.06 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr4_-_150736962 | 0.98 |
ENST00000502345.1
ENST00000510975.1 ENST00000511993.1 |
RP11-526A4.1
|
RP11-526A4.1 |
| chr1_+_246887349 | 0.97 |
ENST00000366510.3
|
SCCPDH
|
saccharopine dehydrogenase (putative) |
| chr7_-_69062391 | 0.95 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr6_+_19837592 | 0.88 |
ENST00000378700.3
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| chr17_-_39140549 | 0.87 |
ENST00000377755.4
|
KRT40
|
keratin 40 |
| chr7_+_138915102 | 0.84 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr4_+_95679072 | 0.83 |
ENST00000515059.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr12_-_91539918 | 0.83 |
ENST00000548218.1
|
DCN
|
decorin |
| chr19_+_38085731 | 0.82 |
ENST00000589117.1
|
ZNF540
|
zinc finger protein 540 |
| chr6_+_127898312 | 0.81 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr4_-_148605265 | 0.81 |
ENST00000541232.1
ENST00000322396.6 |
PRMT10
|
protein arginine methyltransferase 10 (putative) |
| chr6_-_46922659 | 0.79 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chr18_-_10748498 | 0.79 |
ENST00000579949.1
|
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr14_-_68000442 | 0.78 |
ENST00000554278.1
|
TMEM229B
|
transmembrane protein 229B |
| chr6_-_33041378 | 0.75 |
ENST00000428995.1
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr1_+_171107241 | 0.75 |
ENST00000236166.3
|
FMO6P
|
flavin containing monooxygenase 6 pseudogene |
| chr1_+_47533160 | 0.72 |
ENST00000334194.3
|
CYP4Z1
|
cytochrome P450, family 4, subfamily Z, polypeptide 1 |
| chr5_-_41261540 | 0.72 |
ENST00000263413.3
|
C6
|
complement component 6 |
| chr7_-_92777606 | 0.70 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr2_+_89986318 | 0.65 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr6_-_76782371 | 0.65 |
ENST00000369950.3
ENST00000369963.3 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr18_-_74839891 | 0.65 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr17_+_3118915 | 0.64 |
ENST00000304094.1
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1 |
| chr1_-_149832704 | 0.64 |
ENST00000392933.1
ENST00000369157.2 ENST00000392932.4 |
HIST2H4B
|
histone cluster 2, H4b |
| chr5_+_140248518 | 0.63 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr3_+_129159039 | 0.62 |
ENST00000507564.1
ENST00000431818.2 ENST00000504021.1 ENST00000349441.2 ENST00000348417.2 ENST00000440957.2 |
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr5_+_140180635 | 0.61 |
ENST00000522353.2
ENST00000532566.2 |
PCDHA3
|
protocadherin alpha 3 |
| chrX_-_38186775 | 0.61 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr12_-_89413456 | 0.61 |
ENST00000500381.2
|
RP11-13A1.1
|
RP11-13A1.1 |
| chr1_+_43766642 | 0.59 |
ENST00000372476.3
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr9_+_137979506 | 0.57 |
ENST00000539529.1
ENST00000392991.4 ENST00000371793.3 |
OLFM1
|
olfactomedin 1 |
| chr11_+_118175132 | 0.57 |
ENST00000361763.4
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr15_-_78538025 | 0.57 |
ENST00000560817.1
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr11_+_101785727 | 0.57 |
ENST00000263468.8
|
KIAA1377
|
KIAA1377 |
| chr6_-_112080256 | 0.56 |
ENST00000462856.2
ENST00000229471.4 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr17_+_17685422 | 0.56 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr8_+_101170563 | 0.56 |
ENST00000520508.1
ENST00000388798.2 |
SPAG1
|
sperm associated antigen 1 |
| chr10_-_13544945 | 0.55 |
ENST00000378605.3
ENST00000341083.3 |
BEND7
|
BEN domain containing 7 |
| chr1_+_149804218 | 0.52 |
ENST00000610125.1
|
HIST2H4A
|
histone cluster 2, H4a |
| chr6_-_49681235 | 0.52 |
ENST00000339139.4
|
CRISP2
|
cysteine-rich secretory protein 2 |
| chr6_-_11807277 | 0.51 |
ENST00000379415.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr6_-_32784687 | 0.50 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr15_-_76304731 | 0.48 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr1_+_166958346 | 0.48 |
ENST00000367872.4
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr1_-_198990166 | 0.48 |
ENST00000427439.1
|
RP11-16L9.3
|
RP11-16L9.3 |
| chr9_-_88715044 | 0.48 |
ENST00000388711.3
ENST00000466178.1 |
GOLM1
|
golgi membrane protein 1 |
| chr5_-_79287060 | 0.47 |
ENST00000512560.1
ENST00000509852.1 ENST00000512528.1 |
MTX3
|
metaxin 3 |
| chr6_-_52628271 | 0.47 |
ENST00000493422.1
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr9_-_32526299 | 0.46 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr4_+_70894130 | 0.45 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr17_-_10707410 | 0.45 |
ENST00000581851.1
|
LINC00675
|
long intergenic non-protein coding RNA 675 |
| chr12_-_13248598 | 0.43 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr7_+_6797288 | 0.42 |
ENST00000433859.2
ENST00000359718.3 |
RSPH10B2
|
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chr1_+_174844645 | 0.42 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr4_+_70146217 | 0.42 |
ENST00000335568.5
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chr9_-_139137648 | 0.42 |
ENST00000358701.5
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr6_-_130031358 | 0.41 |
ENST00000368149.2
|
ARHGAP18
|
Rho GTPase activating protein 18 |
| chr8_+_118147498 | 0.40 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr17_-_28257080 | 0.40 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr17_-_2415169 | 0.39 |
ENST00000263092.6
ENST00000538844.1 ENST00000576976.1 |
METTL16
|
methyltransferase like 16 |
| chr3_-_123339343 | 0.39 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr3_-_123339418 | 0.38 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr5_-_180076580 | 0.38 |
ENST00000502649.1
|
FLT4
|
fms-related tyrosine kinase 4 |
| chr9_+_86238016 | 0.38 |
ENST00000530832.1
ENST00000405990.3 ENST00000376417.4 ENST00000376419.4 |
IDNK
|
idnK, gluconokinase homolog (E. coli) |
| chr17_+_35732955 | 0.38 |
ENST00000300618.4
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr12_-_9360966 | 0.37 |
ENST00000261336.2
|
PZP
|
pregnancy-zone protein |
| chr1_+_179050512 | 0.37 |
ENST00000367627.3
|
TOR3A
|
torsin family 3, member A |
| chr1_+_174128639 | 0.36 |
ENST00000251507.4
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr3_+_148508845 | 0.36 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr2_+_61372226 | 0.36 |
ENST00000426997.1
|
C2orf74
|
chromosome 2 open reading frame 74 |
| chr5_+_110409012 | 0.36 |
ENST00000379706.4
|
TSLP
|
thymic stromal lymphopoietin |
| chr7_-_152373216 | 0.35 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr3_+_148545586 | 0.34 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr9_+_21967137 | 0.34 |
ENST00000441769.2
|
C9orf53
|
chromosome 9 open reading frame 53 |
| chr9_-_88714421 | 0.34 |
ENST00000388712.3
|
GOLM1
|
golgi membrane protein 1 |
| chr7_+_80275621 | 0.34 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr10_-_113943447 | 0.34 |
ENST00000369425.1
ENST00000348367.4 ENST00000423155.1 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr5_+_101569696 | 0.33 |
ENST00000597120.1
|
AC008948.1
|
AC008948.1 |
| chr1_+_174128536 | 0.33 |
ENST00000357444.6
ENST00000367689.3 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr7_+_80275752 | 0.33 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr15_-_44069741 | 0.32 |
ENST00000319359.3
|
ELL3
|
elongation factor RNA polymerase II-like 3 |
| chr14_-_24911868 | 0.32 |
ENST00000554698.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr19_-_35719609 | 0.32 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187, member B |
| chr17_+_35732916 | 0.31 |
ENST00000586700.1
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr13_+_34392200 | 0.31 |
ENST00000434425.1
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr18_-_24722995 | 0.31 |
ENST00000581714.1
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr3_-_195310802 | 0.31 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr5_-_74532696 | 0.31 |
ENST00000506364.2
ENST00000274361.3 |
ANKRD31
|
ankyrin repeat domain 31 |
| chr8_-_95274536 | 0.30 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr6_+_88182643 | 0.30 |
ENST00000369556.3
ENST00000544441.1 ENST00000369552.4 ENST00000369557.5 |
SLC35A1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1 |
| chr15_-_58571445 | 0.30 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr7_+_80275953 | 0.29 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr11_+_49050504 | 0.29 |
ENST00000332682.7
|
TRIM49B
|
tripartite motif containing 49B |
| chr8_-_10512569 | 0.29 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr11_+_30252549 | 0.28 |
ENST00000254122.3
ENST00000417547.1 |
FSHB
|
follicle stimulating hormone, beta polypeptide |
| chr10_+_118187379 | 0.28 |
ENST00000369230.3
|
PNLIPRP3
|
pancreatic lipase-related protein 3 |
| chr2_-_177502254 | 0.28 |
ENST00000339037.3
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr8_+_77318769 | 0.27 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr9_-_74675521 | 0.27 |
ENST00000377024.3
|
C9orf57
|
chromosome 9 open reading frame 57 |
| chr1_-_112106556 | 0.27 |
ENST00000443498.1
|
ADORA3
|
adenosine A3 receptor |
| chr3_-_51909600 | 0.26 |
ENST00000446461.1
|
IQCF5
|
IQ motif containing F5 |
| chr19_-_11456905 | 0.26 |
ENST00000588560.1
ENST00000592952.1 |
TMEM205
|
transmembrane protein 205 |
| chr1_-_86174065 | 0.25 |
ENST00000370574.3
ENST00000431532.2 |
ZNHIT6
|
zinc finger, HIT-type containing 6 |
| chr17_-_46806540 | 0.25 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chr19_-_11456872 | 0.25 |
ENST00000586218.1
|
TMEM205
|
transmembrane protein 205 |
| chr16_-_2379688 | 0.25 |
ENST00000567910.1
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3 |
| chr19_+_7011509 | 0.25 |
ENST00000377296.3
|
AC025278.1
|
Uncharacterized protein |
| chr19_+_3762645 | 0.25 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr16_+_16481598 | 0.24 |
ENST00000327792.5
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr19_-_11457082 | 0.24 |
ENST00000587948.1
|
TMEM205
|
transmembrane protein 205 |
| chr19_+_38085768 | 0.24 |
ENST00000316433.4
ENST00000590588.1 ENST00000586134.1 ENST00000586792.1 |
ZNF540
|
zinc finger protein 540 |
| chr19_-_11456722 | 0.24 |
ENST00000354882.5
|
TMEM205
|
transmembrane protein 205 |
| chr19_-_11457162 | 0.24 |
ENST00000590482.1
|
TMEM205
|
transmembrane protein 205 |
| chr17_+_29664830 | 0.24 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr4_+_170581213 | 0.24 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr11_-_104480019 | 0.24 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr12_-_4647606 | 0.23 |
ENST00000261250.3
ENST00000541014.1 ENST00000545746.1 ENST00000542080.1 |
C12orf4
|
chromosome 12 open reading frame 4 |
| chr6_+_20534672 | 0.22 |
ENST00000274695.4
ENST00000378624.4 |
CDKAL1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr5_-_58571935 | 0.22 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr22_+_23101182 | 0.21 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr14_-_50065882 | 0.21 |
ENST00000539688.1
|
AL139099.1
|
Full-length cDNA clone CS0DK012YO09 of HeLa cells of Homo sapiens (human); Uncharacterized protein |
| chr8_+_95565947 | 0.21 |
ENST00000523011.1
|
RP11-267M23.4
|
RP11-267M23.4 |
| chrX_+_139791917 | 0.20 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr19_-_38085633 | 0.20 |
ENST00000593133.1
ENST00000590751.1 ENST00000358744.3 ENST00000328550.2 ENST00000451802.2 |
ZNF571
|
zinc finger protein 571 |
| chr14_-_106963409 | 0.20 |
ENST00000390621.2
|
IGHV1-45
|
immunoglobulin heavy variable 1-45 |
| chrX_-_135338503 | 0.20 |
ENST00000370663.5
|
MAP7D3
|
MAP7 domain containing 3 |
| chr5_+_5140436 | 0.20 |
ENST00000511368.1
ENST00000274181.7 |
ADAMTS16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16 |
| chr14_-_50319482 | 0.20 |
ENST00000546046.1
ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF
|
nuclear export mediator factor |
| chr1_-_112106578 | 0.20 |
ENST00000369717.4
|
ADORA3
|
adenosine A3 receptor |
| chr18_-_52989525 | 0.20 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr5_+_42756903 | 0.19 |
ENST00000361970.5
ENST00000388827.4 |
CCDC152
|
coiled-coil domain containing 152 |
| chr14_-_50319758 | 0.19 |
ENST00000298310.5
|
NEMF
|
nuclear export mediator factor |
| chr8_+_133879193 | 0.19 |
ENST00000377869.1
ENST00000220616.4 |
TG
|
thyroglobulin |
| chr2_+_103035102 | 0.19 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr3_+_122513642 | 0.19 |
ENST00000261038.5
|
DIRC2
|
disrupted in renal carcinoma 2 |
| chr1_-_85097431 | 0.19 |
ENST00000327308.3
|
C1orf180
|
chromosome 1 open reading frame 180 |
| chr12_-_54689532 | 0.19 |
ENST00000540264.2
ENST00000312156.4 |
NFE2
|
nuclear factor, erythroid 2 |
| chr19_+_16999654 | 0.18 |
ENST00000248076.3
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr2_-_234475380 | 0.18 |
ENST00000443711.2
ENST00000251722.6 |
USP40
|
ubiquitin specific peptidase 40 |
| chr1_-_153123345 | 0.17 |
ENST00000368748.4
|
SPRR2G
|
small proline-rich protein 2G |
| chr13_-_95131923 | 0.17 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr1_-_168464875 | 0.17 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr10_+_18240834 | 0.17 |
ENST00000377371.3
ENST00000539911.1 |
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr12_-_9885888 | 0.17 |
ENST00000327839.3
|
CLECL1
|
C-type lectin-like 1 |
| chr2_-_225811747 | 0.17 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr22_-_19435209 | 0.17 |
ENST00000546308.1
ENST00000541063.1 ENST00000399568.1 ENST00000333059.5 |
HIRA
C22orf39
|
histone cell cycle regulator chromosome 22 open reading frame 39 |
| chr4_+_48485341 | 0.16 |
ENST00000273861.4
|
SLC10A4
|
solute carrier family 10, member 4 |
| chrX_+_133733457 | 0.16 |
ENST00000440614.1
|
RP11-308B5.2
|
RP11-308B5.2 |
| chr19_-_6279932 | 0.16 |
ENST00000252674.7
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| chr9_-_5304432 | 0.16 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr5_-_169725231 | 0.16 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr21_+_43823983 | 0.16 |
ENST00000291535.6
ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr3_-_46037299 | 0.16 |
ENST00000296137.2
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr10_-_48055018 | 0.16 |
ENST00000426610.2
|
ASAH2C
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2C |
| chr6_-_150217195 | 0.15 |
ENST00000532335.1
|
RAET1E
|
retinoic acid early transcript 1E |
| chr7_+_23749767 | 0.15 |
ENST00000355870.3
|
STK31
|
serine/threonine kinase 31 |
| chr7_+_23749945 | 0.15 |
ENST00000354639.3
ENST00000531170.1 ENST00000444333.2 ENST00000428484.1 |
STK31
|
serine/threonine kinase 31 |
| chr5_+_134181625 | 0.15 |
ENST00000394976.3
|
C5orf24
|
chromosome 5 open reading frame 24 |
| chr1_-_89591749 | 0.15 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr21_+_25801041 | 0.15 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr9_-_2844058 | 0.14 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr8_+_1993152 | 0.14 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr8_+_1993173 | 0.14 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr7_-_4877677 | 0.14 |
ENST00000538469.1
|
RADIL
|
Ras association and DIL domains |
| chr6_-_51952418 | 0.14 |
ENST00000371117.3
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr2_+_27498289 | 0.14 |
ENST00000296097.3
ENST00000420191.1 |
DNAJC5G
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma |
| chr17_+_19091325 | 0.13 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr4_+_110749143 | 0.13 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr17_+_43861680 | 0.13 |
ENST00000314537.5
|
CRHR1
|
corticotropin releasing hormone receptor 1 |
| chr1_+_90460661 | 0.13 |
ENST00000340281.4
ENST00000361911.5 ENST00000370447.3 ENST00000455342.2 |
ZNF326
|
zinc finger protein 326 |
| chr15_+_77224045 | 0.12 |
ENST00000320963.5
ENST00000394883.3 |
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr15_+_77223960 | 0.12 |
ENST00000394885.3
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr7_-_64023441 | 0.12 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chrX_+_49687216 | 0.12 |
ENST00000376088.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr16_-_72206034 | 0.12 |
ENST00000537465.1
ENST00000237353.10 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr5_+_150020214 | 0.12 |
ENST00000307662.4
|
SYNPO
|
synaptopodin |
| chr1_+_222886694 | 0.11 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr12_-_52828147 | 0.11 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chr8_-_86290333 | 0.11 |
ENST00000521846.1
ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1
|
carbonic anhydrase I |
| chr20_+_36946029 | 0.11 |
ENST00000417318.1
|
BPI
|
bactericidal/permeability-increasing protein |
| chr11_+_62037622 | 0.11 |
ENST00000227918.2
ENST00000525380.1 |
SCGB2A2
|
secretoglobin, family 2A, member 2 |
| chr10_+_18240814 | 0.11 |
ENST00000377374.4
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr9_+_103947311 | 0.10 |
ENST00000395056.2
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr20_-_52687059 | 0.10 |
ENST00000371435.2
ENST00000395961.3 |
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr2_-_223520770 | 0.10 |
ENST00000536361.1
|
FARSB
|
phenylalanyl-tRNA synthetase, beta subunit |
| chr2_-_86850949 | 0.10 |
ENST00000237455.4
|
RNF103
|
ring finger protein 103 |
| chr14_+_37641012 | 0.10 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chr22_+_39916558 | 0.10 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr3_+_185080908 | 0.10 |
ENST00000265026.3
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.2 | 0.7 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.2 | 0.9 | GO:0061107 | prostate gland stromal morphogenesis(GO:0060741) seminal vesicle development(GO:0061107) |
| 0.2 | 0.8 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 0.5 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.2 | 0.6 | GO:0035720 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 1.0 | GO:0072564 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.4 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.1 | 0.5 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.6 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.6 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 1.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.6 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.1 | 0.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.6 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.4 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.3 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.4 | GO:0060312 | regulation of blood vessel remodeling(GO:0060312) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.6 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.3 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.2 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.8 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.6 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.0 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.6 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.4 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.3 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 1.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.6 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.5 | GO:0050832 | defense response to fungus(GO:0050832) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0030849 | autosome(GO:0030849) |
| 0.1 | 0.3 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.9 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.6 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.8 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.2 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 1.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.9 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.4 | 1.8 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.4 | 1.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.2 | 1.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.4 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.6 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.8 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.3 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 1.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.0 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 2.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |